Paramyxovirus membrane fusion: lessons from the F and HN atomic structures
- PMID: 16364733
- PMCID: PMC7172328
- DOI: 10.1016/j.virol.2005.09.007
Paramyxovirus membrane fusion: lessons from the F and HN atomic structures
Abstract
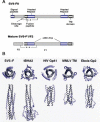
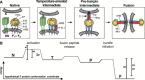
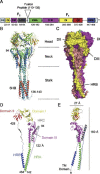
Paramyxoviruses enter cells by fusion of their lipid envelope with the target cell plasma membrane. Fusion of the viral membrane with the plasma membrane allows entry of the viral genome into the cytoplasm. For paramyxoviruses, membrane fusion occurs at neutral pH, but the trigger mechanism that controls the viral entry machinery such that it occurs at the right time and in the right place remains to be elucidated. Two viral glycoproteins are key to the infection process-an attachment protein that varies among different paramyxoviruses and the fusion (F) protein, which is found in all paramyxoviruses. For many of the paramyxoviruses (parainfluenza viruses 1-5, mumps virus, Newcastle disease virus and others), the attachment protein is the hemagglutinin/neuraminidase (HN) protein. In the last 5 years, atomic structures of paramyxovirus F and HN proteins have been reported. The knowledge gained from these structures towards understanding the mechanism of viral membrane fusion is described.
Figures


Similar articles
-
Fusion activation through attachment protein stalk domains indicates a conserved core mechanism of paramyxovirus entry into cells.J Virol. 2014 Apr;88(8):3925-41. doi: 10.1128/JVI.03741-13. Epub 2014 Jan 22. J Virol. 2014. PMID: 24453369 Free PMC article.
-
Interaction between the hemagglutinin-neuraminidase and fusion glycoproteins of human parainfluenza virus type III regulates viral growth in vivo.mBio. 2013 Oct 22;4(5):e00803-13. doi: 10.1128/mBio.00803-13. mBio. 2013. PMID: 24149514 Free PMC article.
-
Premature activation of the paramyxovirus fusion protein before target cell attachment with corruption of the viral fusion machinery.J Biol Chem. 2011 Nov 4;286(44):37945-37954. doi: 10.1074/jbc.M111.256248. Epub 2011 Jul 28. J Biol Chem. 2011. PMID: 21799008 Free PMC article.
-
Structure and function of a paramyxovirus fusion protein.Biochim Biophys Acta. 2003 Jul 11;1614(1):73-84. doi: 10.1016/s0005-2736(03)00164-0. Biochim Biophys Acta. 2003. PMID: 12873767 Review.
-
Activation of paramyxovirus membrane fusion and virus entry.Curr Opin Virol. 2014 Apr;5:24-33. doi: 10.1016/j.coviro.2014.01.005. Epub 2014 Feb 16. Curr Opin Virol. 2014. PMID: 24530984 Free PMC article. Review.
Cited by
-
Novel Functions of Hendra Virus G N-Glycans and Comparisons to Nipah Virus.J Virol. 2015 Jul;89(14):7235-47. doi: 10.1128/JVI.00773-15. Epub 2015 May 6. J Virol. 2015. PMID: 25948743 Free PMC article.
-
In vitro characterization of the antiviral activity of fucoidan from Cladosiphon okamuranus against Newcastle Disease Virus.Virol J. 2012 Dec 12;9:307. doi: 10.1186/1743-422X-9-307. Virol J. 2012. PMID: 23234372 Free PMC article.
-
Spring-loaded heptad repeat residues regulate the expression and activation of paramyxovirus fusion protein.J Virol. 2007 Apr;81(7):3130-41. doi: 10.1128/JVI.02464-06. Epub 2007 Jan 24. J Virol. 2007. PMID: 17251293 Free PMC article.
-
Combinatorial F-G Immunogens as Nipah and Respiratory Syncytial Virus Vaccine Candidates.Viruses. 2021 Sep 28;13(10):1942. doi: 10.3390/v13101942. Viruses. 2021. PMID: 34696372 Free PMC article.
-
Emerging paramyxoviruses: molecular mechanisms and antiviral strategies.Expert Rev Mol Med. 2011 Feb 24;13:e6. doi: 10.1017/S1462399410001754. Expert Rev Mol Med. 2011. PMID: 21345285 Free PMC article. Review.
References
-
- Baker K.A., Dutch R.E., Lamb R.A., Jardetzky T.S. Structural basis for paramyxovirus-mediated membrane fusion. Mol. Cell. 1999;3:309–319. - PubMed
-
- Bullough P.A., Hughson F.M., Skehel J.J., Wiley D.C. Structure of influenza haemagglutinin at the pH of membrane fusion. Nature. 1994;371:37–43. - PubMed
-
- Chan D.C., Fass D., Berger J.M., Kim P.S. Core structure of gp41 from the HIV envelope glycoprotein. Cell. 1997;89:263–273. - PubMed
-
- Chen J., Lee K.H., Steinhauer D.A., Stevens D.J., Skehel J.J., Wiley D.C. Structure of the hemagglutinin precursor cleavage site, a determinant of influenza pathogenicity and the origin of the labile conformation. Cell. 1998;95:409–417. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

