Different domains control the localization and mobility of LIKE HETEROCHROMATIN PROTEIN1 in Arabidopsis nuclei
- PMID: 16361394
- PMCID: PMC1323489
- DOI: 10.1105/tpc.105.036855
Different domains control the localization and mobility of LIKE HETEROCHROMATIN PROTEIN1 in Arabidopsis nuclei
Abstract
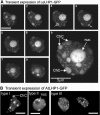
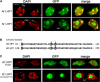
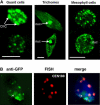
Plants possess a single gene for the structurally related HETEROCHROMATIN PROTEIN1 (HP1), termed LIKE-HP1 (LHP1). We investigated the subnuclear localization, binding properties, and dynamics of LHP1 proteins in Arabidopsis thaliana cells. Transient expression assays showed that tomato (Solanum lycopersicum) LHP1 fused to green fluorescent protein (GFP; Sl LHP1-GFP) and Arabidopsis LHP1 (At LHP1-GFP) localized to heterochromatic chromocenters and showed punctuated distribution within the nucleus; tomato but not Arabidopsis LHP1 was also localized within the nucleolus. Mutations of aromatic cage residues that recognize methyl K9 of histone H3 abolished their punctuated distribution and localization to chromocenters. Sl LHP1-GFP plants displayed cell type-dependent subnuclear localization. The diverse localization pattern of tomato LHP1 did not require the chromo shadow domain (CSD), whereas the chromodomain alone was insufficient for localization to chromocenters; a nucleolar localization signal was identified within the hinge region. Fluorescence recovery after photobleaching showed that Sl LHP1 is a highly mobile protein whose localization and retention are controlled by distinct domains; retention at the nucleolus and chromocenters is conferred by the CSD. Our results imply that LHP1 recruitment to chromatin is mediated, at least in part, through interaction with methyl K9 and that LHP1 controls different nuclear processes via transient binding to its nuclear sites.
Figures
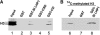
Similar articles
-
The chromodomain of LIKE HETEROCHROMATIN PROTEIN 1 is essential for H3K27me3 binding and function during Arabidopsis development.PLoS One. 2009;4(4):e5335. doi: 10.1371/journal.pone.0005335. Epub 2009 Apr 28. PLoS One. 2009. PMID: 19399177 Free PMC article.
-
The Arabidopsis LHP1 protein is a component of euchromatin.Planta. 2005 Nov;222(5):910-25. doi: 10.1007/s00425-005-0129-4. Epub 2005 Oct 22. Planta. 2005. PMID: 16244868
-
LHP1 Interacts with ATRX through Plant-Specific Domains at Specific Loci Targeted by PRC2.Mol Plant. 2018 Aug 6;11(8):1038-1052. doi: 10.1016/j.molp.2018.05.004. Epub 2018 May 21. Mol Plant. 2018. PMID: 29793052
-
The Arabidopsis GAGA-Binding Factor BASIC PENTACYSTEINE6 Recruits the POLYCOMB-REPRESSIVE COMPLEX1 Component LIKE HETEROCHROMATIN PROTEIN1 to GAGA DNA Motifs.Plant Physiol. 2015 Jul;168(3):1013-24. doi: 10.1104/pp.15.00409. Epub 2015 May 29. Plant Physiol. 2015. PMID: 26025051 Free PMC article.
-
Regulation of heterochromatin organization in plants.J Plant Res. 2024 Sep;137(5):685-693. doi: 10.1007/s10265-024-01550-3. Epub 2024 Jun 24. J Plant Res. 2024. PMID: 38914831 Review.
Cited by
-
Arabidopsis MSI1 connects LHP1 to PRC2 complexes.EMBO J. 2013 Jul 17;32(14):2073-85. doi: 10.1038/emboj.2013.145. Epub 2013 Jun 18. EMBO J. 2013. PMID: 23778966 Free PMC article.
-
LHP1, the Arabidopsis homologue of HETEROCHROMATIN PROTEIN1, is required for epigenetic silencing of FLC.Proc Natl Acad Sci U S A. 2006 Mar 28;103(13):5012-7. doi: 10.1073/pnas.0507427103. Epub 2006 Mar 20. Proc Natl Acad Sci U S A. 2006. PMID: 16549797 Free PMC article.
-
The chromodomain of LIKE HETEROCHROMATIN PROTEIN 1 is essential for H3K27me3 binding and function during Arabidopsis development.PLoS One. 2009;4(4):e5335. doi: 10.1371/journal.pone.0005335. Epub 2009 Apr 28. PLoS One. 2009. PMID: 19399177 Free PMC article.
-
Conservation and divergence of plant LHP1 protein sequences and expression patterns in angiosperms and gymnosperms.Mol Genet Genomics. 2011 May;285(5):357-73. doi: 10.1007/s00438-011-0609-0. Epub 2011 Mar 18. Mol Genet Genomics. 2011. PMID: 21416255
-
The three methyl-CpG-binding domains of AtMBD7 control its subnuclear localization and mobility.J Biol Chem. 2008 Mar 28;283(13):8406-11. doi: 10.1074/jbc.M706221200. Epub 2008 Jan 22. J Biol Chem. 2008. PMID: 18211904 Free PMC article.
References
-
- Andersen, J.S., Lyon, C.E., Fox, A.H., Leung, A.K., Lam, Y.W., Steen, H., Mann, M., and Lamond, A.I. (2002). Directed proteomic analysis of the human nucleolus. Curr. Biol. 12 1–11. - PubMed
-
- Avivi, Y., Morad, V., Ben-Meir, H., Zhao, J., Kashkush, K., Tzfira, T., Citovsky, V., and Grafi, G. (2004). Reorganization of specific chromosomal domains and activation of silent genes in plant cells acquiring pluripotentiality. Dev. Dyn. 230 12–22. - PubMed
-
- Bannister, A.J., Zegerman, P., Partridge, J.F., Miska, E.A., Thomas, J.O., Allshire, R.C., and Kouzarides, T. (2001). Selective recognition of methylated lysine 9 on histone H3 by the HP1 chromo domain. Nature 410 120–124. - PubMed
-
- Brasher, S.V., Smith, B.O., Fogh, R.H., Nietlispach, D., Thiru, A., Nielsen, P.R., Broadhurst, R.W., Ball, L.J., Murzina, N.V., and Laue, E.D. (2000). The structure of mouse HP1 suggests a unique mode of single peptide recognition by the shadow chromo domain dimer. EMBO J. 19 1587–1597. - PMC - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

