Hendra and Nipah viruses: different and dangerous
- PMID: 16357858
- PMCID: PMC7097447
- DOI: 10.1038/nrmicro1323
Hendra and Nipah viruses: different and dangerous
Abstract
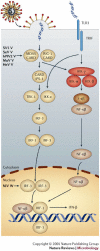
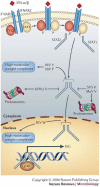
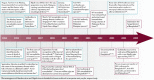
Hendra virus and Nipah virus are highly pathogenic paramyxoviruses that have recently emerged from flying foxes to cause serious disease outbreaks in humans and livestock in Australia, Malaysia, Singapore and Bangladesh. Their unique genetic constitution, high virulence and wide host range set them apart from other paramyxoviruses. These features led to their classification into the new genus Henipavirus within the family Paramyxoviridae and to their designation as Biosafety Level 4 pathogens. This review provides an overview of henipaviruses and the types of infection they cause, and describes how studies on the structure and function of henipavirus proteins expressed from cloned genes have provided insights into the unique biological properties of these emerging human pathogens.
Conflict of interest statement
The authors declare no competing financial interests.
Figures


Similar articles
-
Hendra and Nipah viruses: pathogenesis and therapeutics.Curr Mol Med. 2005 Dec;5(8):805-16. doi: 10.2174/156652405774962308. Curr Mol Med. 2005. PMID: 16375714 Review.
-
Molecular virology of the henipaviruses.Curr Top Microbiol Immunol. 2012;359:41-58. doi: 10.1007/82_2012_211. Curr Top Microbiol Immunol. 2012. PMID: 22552699 Review.
-
A treatment for and vaccine against the deadly Hendra and Nipah viruses.Antiviral Res. 2013 Oct;100(1):8-13. doi: 10.1016/j.antiviral.2013.06.012. Epub 2013 Jul 6. Antiviral Res. 2013. PMID: 23838047 Free PMC article. Review.
-
Hendra and Nipah viruses: why are they so deadly?Curr Opin Virol. 2012 Jun;2(3):242-7. doi: 10.1016/j.coviro.2012.03.006. Epub 2012 Apr 5. Curr Opin Virol. 2012. PMID: 22483665 Review.
-
A review of Nipah and Hendra viruses with an historical aside.Virus Res. 2011 Dec;162(1-2):173-83. doi: 10.1016/j.virusres.2011.09.026. Epub 2011 Sep 22. Virus Res. 2011. PMID: 21963678 Review.
Cited by
-
Nipah virus matrix protein: expert hacker of cellular machines.FEBS Lett. 2016 Aug;590(15):2494-511. doi: 10.1002/1873-3468.12272. Epub 2016 Jul 12. FEBS Lett. 2016. PMID: 27350027 Free PMC article. Review.
-
A Hendra virus G glycoprotein subunit vaccine protects African green monkeys from Nipah virus challenge.Sci Transl Med. 2012 Aug 8;4(146):146ra107. doi: 10.1126/scitranslmed.3004241. Sci Transl Med. 2012. PMID: 22875827 Free PMC article.
-
Role of sequence and structure of the Hendra fusion protein fusion peptide in membrane fusion.J Biol Chem. 2012 Aug 24;287(35):30035-48. doi: 10.1074/jbc.M112.367862. Epub 2012 Jul 3. J Biol Chem. 2012. PMID: 22761418 Free PMC article.
-
Evidence for ubiquitin-regulated nuclear and subnuclear trafficking among Paramyxovirinae matrix proteins.PLoS Pathog. 2015 Mar 17;11(3):e1004739. doi: 10.1371/journal.ppat.1004739. eCollection 2015 Mar. PLoS Pathog. 2015. PMID: 25782006 Free PMC article.
-
An integrated multi-pronged reverse vaccinology and biophysical approaches for identification of potential vaccine candidates against Nipah virus.Saudi Pharm J. 2023 Dec;31(12):101826. doi: 10.1016/j.jsps.2023.101826. Epub 2023 Oct 16. Saudi Pharm J. 2023. PMID: 38028215 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

