Identification of a splicing enhancer in MLH1 using COMPARE, a new assay for determination of relative RNA splicing efficiencies
- PMID: 16357104
- PMCID: PMC1400605
- DOI: 10.1093/hmg/ddi450
Identification of a splicing enhancer in MLH1 using COMPARE, a new assay for determination of relative RNA splicing efficiencies
Abstract
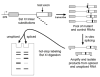
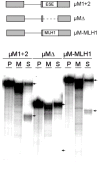
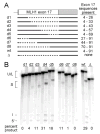
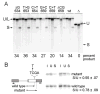
Exonic splicing enhancers (ESEs) are sequences that facilitate recognition of splice sites and prevent exon-skipping. Because ESEs are often embedded within protein-coding sequences, alterations in them can also often be interpreted as nonsense, missense or silent mutations. To correctly interpret exonic mutations and their roles in diseases, it is important to develop strategies that identify ESE mutations. Potential ESEs can be found computationally in many exons but it has proven difficult to predict whether a given mutation will have effects on splicing based on sequence alone. Here, we describe a flexible in vitro method that can be used to functionally compare the effects of multiple sequence variants on ESE activity in a single in vitro splicing reaction. We have applied this method in parallel with conventional splicing assays to test for a splicing enhancer in exon 17 of the human MLH1 gene. Point mutations associated with hereditary non-polyposis colorectal cancer (HNPCC) have previously been found to correlate with exon-skipping in both lymphocytes and tumors from patients. We show that sequences from this exon can replace an ESE from the mouse IgM gene to support RNA splicing in HeLa nuclear extracts. ESE activity was reduced by HNPCC point mutations in codon 659, indicating that their primary effect is on splicing. Surprisingly, the strongest enhancer function mapped to a different region of the exon upstream of this codon. Together, our results indicate that HNPCC point mutations in codon 659 affect an auxillary element that augments the enhancer function to ensure exon inclusion.
Figures


Similar articles
-
Disruption of an exon splicing enhancer in exon 3 of MLH1 is the cause of HNPCC in a Quebec family.J Med Genet. 2006 Feb;43(2):153-6. doi: 10.1136/jmg.2005.031997. Epub 2005 May 27. J Med Genet. 2006. PMID: 15923275 Free PMC article.
-
Systematic mRNA analysis for the effect of MLH1 and MSH2 missense and silent mutations on aberrant splicing.Hum Mutat. 2006 Feb;27(2):145-54. doi: 10.1002/humu.20280. Hum Mutat. 2006. PMID: 16395668
-
Aberrant splicing in MLH1 and MSH2 due to exonic and intronic variants.Hum Genet. 2006 Mar;119(1-2):9-22. doi: 10.1007/s00439-005-0107-8. Epub 2005 Dec 8. Hum Genet. 2006. PMID: 16341550
-
Listening to silence and understanding nonsense: exonic mutations that affect splicing.Nat Rev Genet. 2002 Apr;3(4):285-98. doi: 10.1038/nrg775. Nat Rev Genet. 2002. PMID: 11967553 Review.
-
Exonic splicing enhancers: mechanism of action, diversity and role in human genetic diseases.Trends Biochem Sci. 2000 Mar;25(3):106-10. doi: 10.1016/s0968-0004(00)01549-8. Trends Biochem Sci. 2000. PMID: 10694877 Review.
Cited by
-
Genetic and Clinical Analyses of the KIZ-c.226C>T Variant Resulting in a Dual Mutational Mechanism.Genes (Basel). 2024 Jun 18;15(6):804. doi: 10.3390/genes15060804. Genes (Basel). 2024. PMID: 38927740 Free PMC article.
-
In silico and in vivo splicing analysis of MLH1 and MSH2 missense mutations shows exon- and tissue-specific effects.BMC Genomics. 2006 Sep 22;7:243. doi: 10.1186/1471-2164-7-243. BMC Genomics. 2006. PMID: 16995940 Free PMC article.

