Biased alternative polyadenylation in human tissues
- PMID: 16356263
- PMCID: PMC1414089
- DOI: 10.1186/gb-2005-6-12-r100
Biased alternative polyadenylation in human tissues
Abstract
Background: Alternative polyadenylation is one of the mechanisms in human cells that give rise to a variety of transcripts from a single gene. More than half of the human genes have multiple polyadenylation sites (poly(A) sites), leading to variable mRNA and protein products. Previous studies of individual genes have indicated that alternative polyadenylation could occur in a tissue-specific manner.
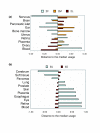
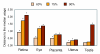
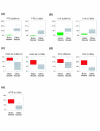
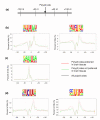
Results: We set out to systematically investigate the occurrence and mechanism of alternative polyadenylation in different human tissues using bioinformatic approaches. Using expressed sequence tag (EST) data, we investigated 42 distinct tissue types. We found that several tissues tend to use poly(A) sites that are biased toward certain locations of a gene, such as sites located in introns or internal exons, and various sites in the exon located closest to the 3' end. We also identified several tissues, including eye, retina and placenta, that tend to use poly(A) sites not frequently used in other tissues. By exploring microarray expression data, we analyzed over 20 genes whose protein products are involved in the process or regulation of mRNA polyadenylation. Several brain tissues showed high concordance of gene expression of these genes with each other, but low concordance with other tissue types. By comparing genomic regions surrounding poly(A) sites preferentially used in brain tissues with those in other tissues, we identified several cis-regulatory elements that were significantly associated with brain-specific poly(A) sites.
Conclusion: Our results indicate that there are systematic differences in poly(A) site usage among human tissues, and both trans-acting factors and cis-regulatory elements may be involved in regulating alternative polyadenylation in different tissues.
Figures

Similar articles
-
Bioinformatic identification of candidate cis-regulatory elements involved in human mRNA polyadenylation.RNA. 2005 Oct;11(10):1485-93. doi: 10.1261/rna.2107305. Epub 2005 Aug 30. RNA. 2005. PMID: 16131587 Free PMC article.
-
A large-scale analysis of mRNA polyadenylation of human and mouse genes.Nucleic Acids Res. 2005 Jan 12;33(1):201-12. doi: 10.1093/nar/gki158. Print 2005. Nucleic Acids Res. 2005. PMID: 15647503 Free PMC article.
-
An intronic polyadenylation site in human and mouse CstF-77 genes suggests an evolutionarily conserved regulatory mechanism.Gene. 2006 Feb 1;366(2):325-34. doi: 10.1016/j.gene.2005.09.024. Epub 2005 Nov 28. Gene. 2006. PMID: 16316725
-
Immunoglobulin heavy chain gene regulation through polyadenylation and splicing competition.Wiley Interdiscip Rev RNA. 2011 Jan-Feb;2(1):92-105. doi: 10.1002/wrna.36. Wiley Interdiscip Rev RNA. 2011. PMID: 21956971 Review.
-
Genome-wide identification and predictive modeling of polyadenylation sites in eukaryotes.Brief Bioinform. 2015 Mar;16(2):304-13. doi: 10.1093/bib/bbu011. Epub 2014 Apr 1. Brief Bioinform. 2015. PMID: 24695098 Review.
Cited by
-
Extensive alternative polyadenylation during zebrafish development.Genome Res. 2012 Oct;22(10):2054-66. doi: 10.1101/gr.139733.112. Epub 2012 Jun 21. Genome Res. 2012. PMID: 22722342 Free PMC article.
-
Intronic cleavage and polyadenylation regulates gene expression during DNA damage response through U1 snRNA.Cell Discov. 2016 Jun 14;2:16013. doi: 10.1038/celldisc.2016.13. eCollection 2016. Cell Discov. 2016. PMID: 27462460 Free PMC article.
-
Analysis of alternative cleavage and polyadenylation by 3' region extraction and deep sequencing.Nat Methods. 2013 Feb;10(2):133-9. doi: 10.1038/nmeth.2288. Epub 2012 Dec 16. Nat Methods. 2013. PMID: 23241633 Free PMC article.
-
Prediction of alternative isoforms from exon expression levels in RNA-Seq experiments.Nucleic Acids Res. 2010 Jun;38(10):e112. doi: 10.1093/nar/gkq041. Epub 2010 Feb 11. Nucleic Acids Res. 2010. PMID: 20150413 Free PMC article.
-
CFIm-mediated alternative polyadenylation remodels cellular signaling and miRNA biogenesis.Nucleic Acids Res. 2022 Apr 8;50(6):3096-3114. doi: 10.1093/nar/gkac114. Nucleic Acids Res. 2022. PMID: 35234914 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

