Serial analysis of gene expression reveals conserved links between protein kinase A, ribosome biogenesis, and phosphate metabolism in Ustilago maydis
- PMID: 16339721
- PMCID: PMC1317500
- DOI: 10.1128/EC.4.12.2029-2043.2005
Serial analysis of gene expression reveals conserved links between protein kinase A, ribosome biogenesis, and phosphate metabolism in Ustilago maydis
Abstract
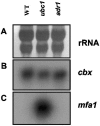
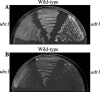
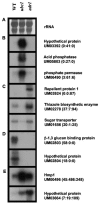
The switch from budding to filamentous growth is a key aspect of invasive growth and virulence for the fungal phytopathogen Ustilago maydis. The cyclic AMP (cAMP) signaling pathway regulates dimorphism in U. maydis, as demonstrated by the phenotypes of mutants with defects in protein kinase A (PKA). Specifically, a mutant lacking the regulatory subunit of PKA encoded by the ubc1 gene displays a multiple-budded phenotype and fails to incite disease symptoms, although proliferation does occur in the plant host. A mutant with a defect in a catalytic subunit of PKA, encoded by adr1, has a constitutively filamentous phenotype and is nonpathogenic. We employed serial analysis of gene expression to examine the transcriptomes of a wild-type strain and the ubc1 and adr1 mutants to further define the role of PKA in U. maydis. The mutants displayed changes in the transcript levels for genes encoding ribosomal proteins, genes regulated by the b mating-type proteins, and genes for metabolic functions. Importantly, the ubc1 mutant displayed elevated transcript levels for genes involved in phosphate acquisition and storage, thus revealing a connection between cAMP and phosphate metabolism. Further experimentation indicated a phosphate storage defect and elevated acid phosphatase activity for the ubc1 mutant. Elevated phosphate levels in culture media also enhanced the filamentous growth of wild-type cells in response to lipids, a finding consistent with PKA regulation of morphogenesis in U. maydis. Overall, these findings extend our understanding of cAMP signaling in U. maydis and reveal a link between phosphate metabolism and morphogenesis.
Figures


Similar articles
-
An Ustilago maydis septin is required for filamentous growth in culture and for full symptom development on maize.Eukaryot Cell. 2005 Dec;4(12):2044-56. doi: 10.1128/EC.4.12.2044-2056.2005. Eukaryot Cell. 2005. PMID: 16339722 Free PMC article.
-
cAMP regulates morphogenesis in the fungal pathogen Ustilago maydis.Genes Dev. 1994 Dec 1;8(23):2805-16. doi: 10.1101/gad.8.23.2805. Genes Dev. 1994. PMID: 7995519
-
Ustilago maydis phosphodiesterases play a role in the dimorphic switch and in pathogenicity.Microbiology (Reading). 2013 May;159(Pt 5):857-868. doi: 10.1099/mic.0.061234-0. Epub 2013 Mar 8. Microbiology (Reading). 2013. PMID: 23475947
-
Genetics of morphogenesis and pathogenic development of Ustilago maydis.Adv Genet. 2007;57:1-47. doi: 10.1016/S0065-2660(06)57001-4. Adv Genet. 2007. PMID: 17352901 Review.
-
Sending mixed signals: redundancy vs. uniqueness of signaling components in the plant pathogen, Ustilago maydis.Fungal Genet Biol. 2008 Aug;45 Suppl 1:S22-30. doi: 10.1016/j.fgb.2008.04.007. Epub 2008 May 23. Fungal Genet Biol. 2008. PMID: 18502157 Review.
Cited by
-
Assessing the impact of transcriptomics, proteomics and metabolomics on fungal phytopathology.Mol Plant Pathol. 2009 Sep;10(5):703-15. doi: 10.1111/j.1364-3703.2009.00565.x. Mol Plant Pathol. 2009. PMID: 19694958 Free PMC article. Review.
-
The emerging role of septins in fungal pathogenesis.Cytoskeleton (Hoboken). 2023 Jul-Aug;80(7-8):242-253. doi: 10.1002/cm.21765. Epub 2023 Jun 2. Cytoskeleton (Hoboken). 2023. PMID: 37265147 Free PMC article. Review.
-
Comparative transcriptome analysis of two Cercospora sojina strains reveals differences in virulence under nitrogen starvation stress.BMC Microbiol. 2020 Jun 16;20(1):166. doi: 10.1186/s12866-020-01853-0. BMC Microbiol. 2020. PMID: 32546122 Free PMC article.
-
An Ustilago maydis septin is required for filamentous growth in culture and for full symptom development on maize.Eukaryot Cell. 2005 Dec;4(12):2044-56. doi: 10.1128/EC.4.12.2044-2056.2005. Eukaryot Cell. 2005. PMID: 16339722 Free PMC article.
-
Two phosphodiesterase genes, PDEL and PDEH, regulate development and pathogenicity by modulating intracellular cyclic AMP levels in Magnaporthe oryzae.PLoS One. 2011 Feb 28;6(2):e17241. doi: 10.1371/journal.pone.0017241. PLoS One. 2011. PMID: 21386978 Free PMC article.
References
-
- Andrews, D. L., J. D. Egan, M. E. Mayorga, and S. E. Gold. 2000. The Ustilago maydis ubc4 and ubc5 genes encode members of a MAP kinase cascade required for filamentous growth. Mol. Plant-Microbe Interact. 13:781-786. - PubMed
-
- Andrews, D. L., M. D. Garcia-Pedrajas, and S. E. Gold. 2004. Fungal dimorphism regulated gene expression in Ustilago maydis: I. Filament up-regulated genes. Mol. Plant Pathol. 5:281-293. - PubMed
-
- Audic, S., and J. M. Claverie. 1997. The significance of digital gene expression profiles. Genome Res. 7:986-995. - PubMed
-
- Banuett, F., and I. Herskowitz. 1994. Identification of fuz7, a Ustilago maydis MEK/MAPKK homolog required for a-locus-dependent and -independent steps in the fungal life cycle. Genes Dev. 8:1367-1378. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

