PGC-1alpha coactivates PDK4 gene expression via the orphan nuclear receptor ERRalpha: a mechanism for transcriptional control of muscle glucose metabolism
- PMID: 16314495
- PMCID: PMC1316952
- DOI: 10.1128/MCB.25.24.10684-10694.2005
PGC-1alpha coactivates PDK4 gene expression via the orphan nuclear receptor ERRalpha: a mechanism for transcriptional control of muscle glucose metabolism
Abstract
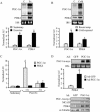
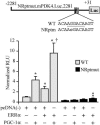
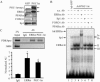
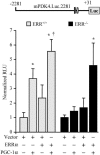
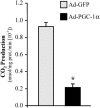
The transcriptional coactivator PGC-1alpha is a key regulator of energy metabolism, yet little is known about its role in control of substrate selection. We found that physiological stimuli known to induce PGC-1alpha expression in skeletal muscle coordinately upregulate the expression of pyruvate dehydrogenase kinase 4 (PDK4), a negative regulator of glucose oxidation. Forced expression of PGC-1alpha in C(2)C(12) myotubes induced PDK4 mRNA and protein expression. PGC-1alpha-mediated activation of PDK4 expression was shown to occur at the transcriptional level and was mapped to a putative nuclear receptor binding site. Gel shift assays demonstrated that the PGC-1alpha-responsive element bound the estrogen-related receptor alpha (ERRalpha), a recently identified component of the PGC-1alpha signaling pathway. In addition, PGC-1alpha was shown to activate ERRalpha expression. Chromatin immunoprecipitation assays confirmed that PGC-1alpha and ERRalpha occupied the mPDK4 promoter in C(2)C(12) myotubes. Additionally, transfection studies using ERRalpha-null primary fibroblasts demonstrated that ERRalpha is required for PGC-1alpha-mediated activation of the mPDK4 promoter. As predicted by the effects of PGC-1alpha on PDK4 gene transcription, overexpression of PGC-1alpha in C(2)C(12) myotubes decreased glucose oxidation rates. These results identify the PDK4 gene as a new PGC-1alpha/ERRalpha target and suggest a mechanism whereby PGC-1alpha exerts reciprocal inhibitory influences on glucose catabolism while increasing alternate mitochondrial oxidative pathways in skeletal muscle.
Figures
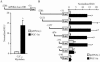


Similar articles
-
Estrogen-related receptor alpha directs peroxisome proliferator-activated receptor alpha signaling in the transcriptional control of energy metabolism in cardiac and skeletal muscle.Mol Cell Biol. 2004 Oct;24(20):9079-91. doi: 10.1128/MCB.24.20.9079-9091.2004. Mol Cell Biol. 2004. PMID: 15456881 Free PMC article.
-
Identification of ERRalpha as a specific partner of PGC-1alpha for the activation of PDK4 gene expression in muscle.FEBS J. 2006 Apr;273(8):1669-80. doi: 10.1111/j.1742-4658.2006.05183.x. FEBS J. 2006. PMID: 16623704
-
Estrogen-related receptor-gamma and peroxisome proliferator-activated receptor-gamma coactivator-1alpha regulate estrogen-related receptor-alpha gene expression via a conserved multi-hormone response element.J Mol Endocrinol. 2005 Apr;34(2):473-87. doi: 10.1677/jme.1.01586. J Mol Endocrinol. 2005. PMID: 15821111
-
[Transcriptional regulation of metabolic switching PDK4 gene under various physiological conditions].Yakugaku Zasshi. 2007 Jan;127(1):153-62. doi: 10.1248/yakushi.127.153. Yakugaku Zasshi. 2007. PMID: 17202796 Review. Japanese.
-
PGC-1alpha-induced improvements in skeletal muscle metabolism and insulin sensitivity.Appl Physiol Nutr Metab. 2009 Jun;34(3):307-14. doi: 10.1139/H09-008. Appl Physiol Nutr Metab. 2009. PMID: 19448691 Review.
Cited by
-
MicroRNA Transcriptome Profile Analysis in Porcine Muscle and the Effect of miR-143 on the MYH7 Gene and Protein.PLoS One. 2015 Apr 27;10(4):e0124873. doi: 10.1371/journal.pone.0124873. eCollection 2015. PLoS One. 2015. PMID: 25915937 Free PMC article.
-
Organismal carbohydrate and lipid homeostasis.Cold Spring Harb Perspect Biol. 2012 May 1;4(5):a006031. doi: 10.1101/cshperspect.a006031. Cold Spring Harb Perspect Biol. 2012. PMID: 22550228 Free PMC article. Review.
-
Exercise intensity-dependent regulation of peroxisome proliferator-activated receptor coactivator-1 mRNA abundance is associated with differential activation of upstream signalling kinases in human skeletal muscle.J Physiol. 2010 May 15;588(Pt 10):1779-90. doi: 10.1113/jphysiol.2010.188011. Epub 2010 Mar 22. J Physiol. 2010. PMID: 20308248 Free PMC article.
-
PGC-1β regulates angiogenesis in skeletal muscle.Am J Physiol Endocrinol Metab. 2011 Jul;301(1):E155-63. doi: 10.1152/ajpendo.00681.2010. Epub 2011 Mar 1. Am J Physiol Endocrinol Metab. 2011. PMID: 21364124 Free PMC article.
-
Involvement of estrogen-related receptors in transcriptional response to hypoxia and growth of solid tumors.Proc Natl Acad Sci U S A. 2008 Jun 3;105(22):7821-6. doi: 10.1073/pnas.0711677105. Epub 2008 May 28. Proc Natl Acad Sci U S A. 2008. PMID: 18509053 Free PMC article.
References
-
- Arany, Z., H. He, J. Lin, K. Hoyer, C. Handschin, O. Toka, F. Ahmad, T. Matsui, S. Chin, P.-H. Wu, I. I. Rybkin, J. M. Shelton, M. Manieri, S. Cinti, F. J. Schoen, R. Bassel-Duby, A. Rosenzweig, J. S. Ingwall, and B. M. Spiegelman. 2005. Transcriptional coactivator PGC-1α controls the energy state and contractile function of cardiac muscle. Cell Metab. 1:259-271. - PubMed
-
- Baar, K., A. R. Wende, T. E. Jones, M. Marison, L. A. Nolte, M. Chen, D. P. Kelly, and J. O. Holloszy. 2002. Adaptations of skeletal muscle to exercise: rapid increase in the transcriptional coactivator PGC-1α. FASEB J. 16:1879-1886. - PubMed
-
- Brandt, J., F. Djouadi, and D. P. Kelly. 1998. Fatty acids activate transcription of the muscle carnitine palmitoyltransferase I gene in cardiac myocytes via the peroxisome proliferator-activated receptor α. J. Biol. Chem. 273:23786-23792. - PubMed
-
- Carter, M. E., T. Gulick, B. D. Raisher, T. Caira, J. A. Ladias, D. D. Moore, and D. P. Kelly. 1993. Hepatocyte nuclear factor-4 activates medium chain acyl-CoA dehydrogenase gene transcription by interacting with a complex regulatory element. J. Biol. Chem. 268:13805-13810. - PubMed
-
- Caruso, M., C. Miele, P. Formisano, G. Condorelli, G. Bifulco, A. Oliva, R. Auricchio, G. Riccardi, B. Capaaldo, and F. Beguinot. 1997. In skeletal muscle, glucose storage and oxidation are differentially impaired by the IR1152 mutant receptor. J. Biol. Chem. 272:7290-7297. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
