Genetic analysis of influenza virus NS1 gene: a temperature-sensitive mutant shows defective formation of virus particles
- PMID: 16306596
- PMCID: PMC1316024
- DOI: 10.1128/JVI.79.24.15246-15257.2005
Genetic analysis of influenza virus NS1 gene: a temperature-sensitive mutant shows defective formation of virus particles
Abstract
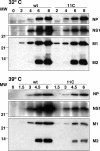
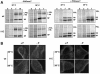
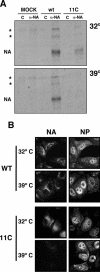
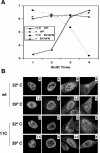
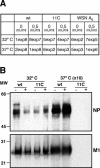
To perform a genetic analysis of the influenza A virus NS1 gene, a library of NS1 mutants was generated by PCR-mediated mutagenesis. A collection of mutant ribonucleic proteins containing the nonstructural genes was generated from the library that were rescued for an infectious virus mutant library by a novel RNP competition virus rescue procedure. Several temperature-sensitive (ts) mutant viruses were obtained by screening of the mutant library, and the sequences of their NS1 genes were determined. Most of the mutations identified led to amino acid exchanges and concentrated in the N-terminal region of the protein, but some of them occurred in the C-terminal region. Mutant 11C contained three mutations that led to amino acid exchanges, V18A, R44K, and S195P, all of which were required for the ts phenotype, and was characterized further. Several steps in the infection were slightly altered: (i) M1, M2, NS1, and neuraminidase (NA) accumulations were reduced and (ii) NS1 protein was retained in the nucleus in a temperature-independent manner, but these modifications could not justify the strong virus titer reduction at restrictive temperature. The most dramatic phenotype was the almost complete absence of virus particles in the culture medium, in spite of normal accumulation and nucleocytoplasmic export of virus RNPs. The function affected in the 11C mutant was required late in the infection, as documented by shift-up and shift-down experiments. The defect in virion production was not due to reduced NA expression, as virus yield could not be rescued by exogenous neuraminidase treatment. All together, the analysis of 11C mutant phenotype may indicate a role for NS1 protein in a late event in virus morphogenesis.
Figures






Similar articles
-
Defective RNA replication and late gene expression in temperature-sensitive influenza viruses expressing deleted forms of the NS1 protein.J Virol. 2004 Apr;78(8):3880-8. doi: 10.1128/jvi.78.8.3880-3888.2004. J Virol. 2004. PMID: 15047804 Free PMC article.
-
Characterization of influenza virus NS1 protein by using a novel helper-virus-free reverse genetic system.J Virol. 2000 Jun;74(12):5556-61. doi: 10.1128/jvi.74.12.5556-5561.2000. J Virol. 2000. PMID: 10823862 Free PMC article.
-
An A14U Substitution in the 3' Noncoding Region of the M Segment of Viral RNA Supports Replication of Influenza Virus with an NS1 Deletion by Modulating Alternative Splicing of M Segment mRNAs.J Virol. 2015 Oct;89(20):10273-85. doi: 10.1128/JVI.00919-15. Epub 2015 Jul 29. J Virol. 2015. PMID: 26223635 Free PMC article.
-
Nuclear functions of the influenza A and B viruses NS1 proteins: do they play a role in viral mRNA export?Vaccine. 2009 Oct 23;27(45):6312-6. doi: 10.1016/j.vaccine.2009.01.015. Vaccine. 2009. PMID: 19840666 Review.
-
[RNA synthesis of influenza A viruses--temperature-sensitive mutants defective in RNA synthesis].Uirusu. 1986 Dec;36(2):203-19. doi: 10.2222/jsv.36.203. Uirusu. 1986. PMID: 3551323 Review. Japanese. No abstract available.
Cited by
-
Influenza virus mRNA translation revisited: is the eIF4E cap-binding factor required for viral mRNA translation?J Virol. 2007 Nov;81(22):12427-38. doi: 10.1128/JVI.01105-07. Epub 2007 Sep 12. J Virol. 2007. PMID: 17855553 Free PMC article.
-
Severe acute respiratory syndrome coronavirus accessory proteins 6 and 9b interact in vivo.Virus Res. 2012 Oct;169(1):282-8. doi: 10.1016/j.virusres.2012.07.012. Epub 2012 Jul 20. Virus Res. 2012. PMID: 22820404 Free PMC article.
-
Inhibition of Influenza Virus Replication by DNA Aptamers Targeting a Cellular Component of Translation Initiation.Mol Ther Nucleic Acids. 2016 Apr 12;5(4):e308. doi: 10.1038/mtna.2016.20. Mol Ther Nucleic Acids. 2016. PMID: 27070300 Free PMC article.
-
hCLE/C14orf166, a cellular protein required for viral replication, is incorporated into influenza virus particles.Sci Rep. 2016 Feb 11;6:20744. doi: 10.1038/srep20744. Sci Rep. 2016. PMID: 26864902 Free PMC article.
-
Human Staufen1 protein interacts with influenza virus ribonucleoproteins and is required for efficient virus multiplication.J Virol. 2010 Aug;84(15):7603-12. doi: 10.1128/JVI.00504-10. Epub 2010 May 26. J Virol. 2010. PMID: 20504931 Free PMC article.
References
-
- Blom, N., S. Gammeltoft, and S. Brunak. 1999. Sequence and structure-based prediction of eukaryotic protein phosphorylation sites. J. Mol. Biol. 294:1351-1362. - PubMed
-
- Briedis, D. J., G. Conti, E. A. Munn, and B. W. J. Mahy. 1981. Migration of influenza virus-specific polypeptides from cytoplasm to nucleus of infected cells. Virology 111:154-164. - PubMed
-
- Burgui, I., T. Aragón, J. Ortín, and A. Nieto. 2003. PABP1 and eIF4GI associate to influenza virus NS1 protein in viral mRNA translation initiation complexes. J. Gen. Virol. 84:3263-3274. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

