The DHHC protein Pfa3 affects vacuole-associated palmitoylation of the fusion factor Vac8
- PMID: 16301533
- PMCID: PMC1297695
- DOI: 10.1073/pnas.0508885102
The DHHC protein Pfa3 affects vacuole-associated palmitoylation of the fusion factor Vac8
Abstract
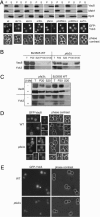
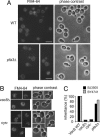
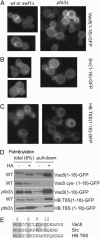
Vacuole biogenesis depends on specific targeting and retention of peripheral membrane proteins. At least three palmitoylated proteins are found exclusively on yeast vacuoles: the fusion factor Vac8, the kinase Yck3, and a novel adaptor protein implicated in microautophagy, Meh1. Here, we analyze the role that putative acyltransferases of the DHHC family play in their localization and function. We find that Pfa3/Ynl326c is required for efficient localization of Vac8 to vacuoles in vivo, while Yck3 or Meh1 localization is not impaired in any of the seven DHHC deletions. Vacuole-associated Vac8 appears to be palmitoylated in a pfa3 mutant, but this population is refractive to further palmitoylation on isolated vacuoles. Vacuole morphology and inheritance, which both depend on Vac8 palmitoylation, appear normal, although there is a reduction in vacuole fusion. Interestingly, Pfa3 is required for the vacuolar localization of not only an SH4 domain that is targeted by myristate/palmitate (as in Vac8) but also one that is targeted by a myristate/basic stretch (as in Src). Our data indicate that Pfa3 has an important but not exclusive function for Vac8 localization to the vacuole.
Figures
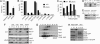
Similar articles
-
Molecular recognition of the palmitoylation substrate Vac8 by its palmitoyltransferase Pfa3.J Biol Chem. 2009 Jun 26;284(26):17720-30. doi: 10.1074/jbc.M109.005447. Epub 2009 May 5. J Biol Chem. 2009. PMID: 19416974 Free PMC article.
-
Palmitoylation determines the function of Vac8 at the yeast vacuole.J Cell Sci. 2006 Jun 15;119(Pt 12):2477-85. doi: 10.1242/jcs.02972. Epub 2006 May 23. J Cell Sci. 2006. PMID: 16720644
-
Probing protein palmitoylation at the yeast vacuole.Methods. 2006 Oct;40(2):171-6. doi: 10.1016/j.ymeth.2006.06.020. Methods. 2006. PMID: 17012029 Review.
-
Analysis of DHHC acyltransferases implies overlapping substrate specificity and a two-step reaction mechanism.Traffic. 2009 Aug;10(8):1061-73. doi: 10.1111/j.1600-0854.2009.00925.x. Epub 2009 May 12. Traffic. 2009. PMID: 19453970
-
Protein transport from the late Golgi to the vacuole in the yeast Saccharomyces cerevisiae.Biochim Biophys Acta. 2005 Jul 10;1744(3):438-54. doi: 10.1016/j.bbamcr.2005.04.004. Biochim Biophys Acta. 2005. PMID: 15913810 Review.
Cited by
-
A novel subtype of AP-1-binding motif within the palmitoylated trans-Golgi network/endosomal accessory protein Gadkin/gamma-BAR.J Biol Chem. 2010 Feb 5;285(6):4074-4086. doi: 10.1074/jbc.M109.049197. Epub 2009 Dec 3. J Biol Chem. 2010. PMID: 19965873 Free PMC article.
-
Discovery of protein-palmitoylating enzymes.Pflugers Arch. 2008 Sep;456(6):1199-206. doi: 10.1007/s00424-008-0465-x. Epub 2008 Jan 30. Pflugers Arch. 2008. PMID: 18231805 Review.
-
2-Bromopalmitate and 2-(2-hydroxy-5-nitro-benzylidene)-benzo[b]thiophen-3-one inhibit DHHC-mediated palmitoylation in vitro.J Lipid Res. 2009 Feb;50(2):233-42. doi: 10.1194/jlr.M800270-JLR200. Epub 2008 Sep 30. J Lipid Res. 2009. PMID: 18827284 Free PMC article.
-
Spatial organization of palmitoyl acyl transferases governs substrate localization and function.Mol Membr Biol. 2019 Dec;35(1):60-75. doi: 10.1080/09687688.2019.1710274. Mol Membr Biol. 2019. PMID: 31969037 Free PMC article. Review.
-
Target of rapamycin (TOR) in nutrient signaling and growth control.Genetics. 2011 Dec;189(4):1177-201. doi: 10.1534/genetics.111.133363. Genetics. 2011. PMID: 22174183 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

