Slx4 regulates DNA damage checkpoint-dependent phosphorylation of the BRCT domain protein Rtt107/Esc4
- PMID: 16267268
- PMCID: PMC1345688
- DOI: 10.1091/mbc.e05-08-0785
Slx4 regulates DNA damage checkpoint-dependent phosphorylation of the BRCT domain protein Rtt107/Esc4
Abstract
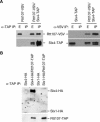
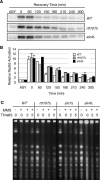
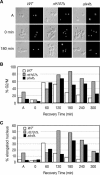
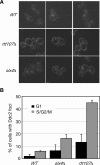
RTT107 (ESC4, YHR154W) encodes a BRCA1 C-terminal-domain protein that is important for recovery from DNA damage during S phase. Rtt107 is a substrate of the checkpoint protein kinase Mec1, although the mechanism by which Rtt107 is targeted by Mec1 after checkpoint activation is currently unclear. Slx4, a component of the Slx1-Slx4 structure-specific nuclease, formed a complex with Rtt107. Deletion of SLX4 conferred many of the same DNA-repair defects observed in rtt107delta, including DNA damage sensitivity, prolonged DNA damage checkpoint activation, and increased spontaneous DNA damage. These phenotypes were not shared by the Slx4 binding partner Slx1, suggesting that the functions of the Slx4 and Slx1 proteins in the DNA damage response were not identical. Of particular interest, Slx4, but not Slx1, was required for phosphorylation of Rtt107 by Mec1 in vivo, indicating that Slx4 was a mediator of DNA damage-dependent phosphorylation of the checkpoint effector Rtt107. We propose that Slx4 has roles in the DNA damage response that are distinct from the function of Slx1-Slx4 in maintaining rDNA structure and that Slx4-dependent phosphorylation of Rtt107 by Mec1 is critical for replication restart after alkylation damage.
Figures



Similar articles
-
Loss of H3 K79 trimethylation leads to suppression of Rtt107-dependent DNA damage sensitivity through the translesion synthesis pathway.J Biol Chem. 2010 Nov 5;285(45):35113-22. doi: 10.1074/jbc.M110.116855. Epub 2010 Sep 1. J Biol Chem. 2010. PMID: 20810656 Free PMC article.
-
DNA damage signaling recruits the Rtt107-Slx4 scaffolds via Dpb11 to mediate replication stress response.Mol Cell. 2010 Jul 30;39(2):300-6. doi: 10.1016/j.molcel.2010.06.019. Mol Cell. 2010. PMID: 20670896
-
Rtt107 BRCT domains act as a targeting module in the DNA damage response.DNA Repair (Amst). 2016 Jan;37:22-32. doi: 10.1016/j.dnarep.2015.10.007. Epub 2015 Nov 10. DNA Repair (Amst). 2016. PMID: 26641499
-
Slx4 scaffolding in homologous recombination and checkpoint control: lessons from yeast.Chromosoma. 2017 Feb;126(1):45-58. doi: 10.1007/s00412-016-0600-y. Epub 2016 May 10. Chromosoma. 2017. PMID: 27165041 Free PMC article. Review.
-
Multi-BRCT scaffolds use distinct strategies to support genome maintenance.Cell Cycle. 2016 Oct;15(19):2561-2570. doi: 10.1080/15384101.2016.1218102. Epub 2016 Aug 11. Cell Cycle. 2016. PMID: 27580271 Free PMC article. Review.
Cited by
-
The Rtt107 BRCT scaffold and its partner modification enzymes collaborate to promote replication.Nucleus. 2016 Jul 3;7(4):346-51. doi: 10.1080/19491034.2016.1201624. Epub 2016 Jul 6. Nucleus. 2016. PMID: 27385431 Free PMC article. Review.
-
Phylogenomic analysis of the GIY-YIG nuclease superfamily.BMC Genomics. 2006 Apr 28;7:98. doi: 10.1186/1471-2164-7-98. BMC Genomics. 2006. PMID: 16646971 Free PMC article.
-
Loss of H3 K79 trimethylation leads to suppression of Rtt107-dependent DNA damage sensitivity through the translesion synthesis pathway.J Biol Chem. 2010 Nov 5;285(45):35113-22. doi: 10.1074/jbc.M110.116855. Epub 2010 Sep 1. J Biol Chem. 2010. PMID: 20810656 Free PMC article.
-
The Saccharomyces cerevisiae Esc2 and Smc5-6 proteins promote sister chromatid junction-mediated intra-S repair.Mol Biol Cell. 2009 Mar;20(6):1671-82. doi: 10.1091/mbc.e08-08-0875. Epub 2009 Jan 21. Mol Biol Cell. 2009. PMID: 19158389 Free PMC article.
-
TOPBP1Dpb11 plays a conserved role in homologous recombination DNA repair through the coordinated recruitment of 53BP1Rad9.J Cell Biol. 2017 Mar 6;216(3):623-639. doi: 10.1083/jcb.201607031. Epub 2017 Feb 22. J Cell Biol. 2017. PMID: 28228534 Free PMC article.
References
-
- Bartkova, J., et al. (2005). DNA damage response as a candidate anti-cancer barrier in early human tumorigenesis. Nature 434, 864-870. - PubMed
-
- Bartrand, A. J., Iyasu, D., and Brush, G. S. (2004). DNA stimulates Mec1-mediated phosphorylation of replication protein A. J. Biol. Chem. 279, 26762-26767. - PubMed
-
- Brachmann, C. B., Davies, A., Cost, G. J., Caputo, E., Li, J., Hieter, P., and Boeke, J. D. (1998). Designer deletion strains derived from Saccharomyces cerevisiae S288C: a useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast 14, 115-132. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

