Spatial and temporal regulation of cofilin activity by LIM kinase and Slingshot is critical for directional cell migration
- PMID: 16230460
- PMCID: PMC2171197
- DOI: 10.1083/jcb.200504029
Spatial and temporal regulation of cofilin activity by LIM kinase and Slingshot is critical for directional cell migration
Abstract
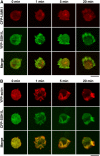
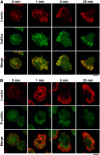
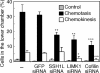
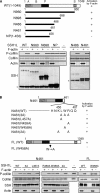
Cofilin mediates lamellipodium extension and polarized cell migration by accelerating actin filament dynamics at the leading edge of migrating cells. Cofilin is inactivated by LIM kinase (LIMK)-1-mediated phosphorylation and is reactivated by cofilin phosphatase Slingshot (SSH)-1L. In this study, we show that cofilin activity is temporally and spatially regulated by LIMK1 and SSH1L in chemokine-stimulated Jurkat T cells. The knockdown of LIMK1 suppressed chemokine-induced lamellipodium formation and cell migration, whereas SSH1L knockdown produced and retained multiple lamellipodial protrusions around the cell after cell stimulation and impaired directional cell migration. Our results indicate that LIMK1 is required for cell migration by stimulating lamellipodium formation in the initial stages of cell response and that SSH1L is crucially involved in directional cell migration by restricting the membrane protrusion to one direction and locally stimulating cofilin activity in the lamellipodium in the front of the migrating cell. We propose that LIMK1- and SSH1L-mediated spatiotemporal regulation of cofilin activity is critical for chemokine-induced polarized lamellipodium formation and directional cell movement.
Figures




Similar articles
-
Critical roles of actin-interacting protein 1 in cytokinesis and chemotactic migration of mammalian cells.Biochem J. 2008 Sep 1;414(2):261-70. doi: 10.1042/BJ20071655. Biochem J. 2008. PMID: 18494608
-
A pathway of neuregulin-induced activation of cofilin-phosphatase Slingshot and cofilin in lamellipodia.J Cell Biol. 2004 May 24;165(4):465-71. doi: 10.1083/jcb.200401136. J Cell Biol. 2004. PMID: 15159416 Free PMC article.
-
Dual regulation of cofilin activity by LIM kinase and Slingshot-1L phosphatase controls platelet-derived growth factor-induced migration of human aortic smooth muscle cells.Circ Res. 2008 Feb 29;102(4):432-8. doi: 10.1161/CIRCRESAHA.107.158923. Epub 2007 Dec 20. Circ Res. 2008. PMID: 18096821
-
Lim kinases, regulators of actin dynamics.Int J Biochem Cell Biol. 2007;39(6):1071-6. doi: 10.1016/j.biocel.2006.11.011. Epub 2006 Nov 28. Int J Biochem Cell Biol. 2007. PMID: 17188549 Review.
-
Cofilin phosphatases and regulation of actin dynamics.Curr Opin Cell Biol. 2006 Feb;18(1):26-31. doi: 10.1016/j.ceb.2005.11.005. Epub 2005 Dec 7. Curr Opin Cell Biol. 2006. PMID: 16337782 Review.
Cited by
-
Direct stimulation of receptor-controlled phospholipase D1 by phospho-cofilin.EMBO J. 2007 Oct 3;26(19):4189-202. doi: 10.1038/sj.emboj.7601852. Epub 2007 Sep 13. EMBO J. 2007. PMID: 17853892 Free PMC article.
-
Slingshot isoform-specific regulation of cofilin-mediated vascular smooth muscle cell migration and neointima formation.Arterioscler Thromb Vasc Biol. 2011 Nov;31(11):2424-31. doi: 10.1161/ATVBAHA.111.232769. Arterioscler Thromb Vasc Biol. 2011. PMID: 21868701 Free PMC article.
-
Chronophin coordinates cell leading edge dynamics by controlling active cofilin levels.Proc Natl Acad Sci U S A. 2015 Sep 15;112(37):E5150-9. doi: 10.1073/pnas.1510945112. Epub 2015 Aug 31. Proc Natl Acad Sci U S A. 2015. PMID: 26324884 Free PMC article.
-
Poly(ADP-ribose) glycohydrolase silencing down-regulates TCTP and Cofilin-1 associated with metastasis in benzo(a)pyrene carcinogenesis.Am J Cancer Res. 2014 Dec 15;5(1):155-67. eCollection 2015. Am J Cancer Res. 2014. PMID: 25628927 Free PMC article.
-
Novel mechanism for negatively regulating Rho-kinase (ROCK) signaling through Coronin1B protein in neuregulin 1 (NRG-1)-induced tumor cell motility.J Biol Chem. 2012 Jun 22;287(26):21836-45. doi: 10.1074/jbc.M112.346114. Epub 2012 May 4. J Biol Chem. 2012. PMID: 22563075 Free PMC article.
References
-
- Arber, S., F.A. Barbayannis, H. Hanser, C. Schneider, C.A. Stanyon, O. Bernard, and P. Caroni. 1998. Regulation of actin dynamics through phosphorylation of cofilin by LIM-kinase. Nature. 393:805–809. - PubMed
-
- Bailly, M., and G.E. Jones. 2003. Polarised migration: cofilin holds the front. Curr. Biol. 13:R128–R130. - PubMed
-
- Bamburg, J.R. 1999. Proteins of the ADF/cofilin family: essential regulators of actin dynamics. Annu. Rev. Cell Dev. Biol. 15:185–230. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

