Human Lsg1 defines a family of essential GTPases that correlates with the evolution of compartmentalization
- PMID: 16209721
- PMCID: PMC1262696
- DOI: 10.1186/1741-7007-3-21
Human Lsg1 defines a family of essential GTPases that correlates with the evolution of compartmentalization
Abstract
Background: Compartmentalization is a key feature of eukaryotic cells, but its evolution remains poorly understood. GTPases are the oldest enzymes that use nucleotides as substrates and they participate in a wide range of cellular processes. Therefore, they are ideal tools for comparative genomic studies aimed at understanding how aspects of biological complexity such as cellular compartmentalization evolved.
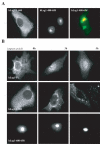
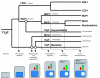
Results: We describe the identification and characterization of a unique family of circularly permuted GTPases represented by the human orthologue of yeast Lsg1p. We placed the members of this family in the phylogenetic context of the YlqF Related GTPase (YRG) family, which are present in Eukarya, Bacteria and Archea and include the stem cell regulator Nucleostemin. To extend the computational analysis, we showed that hLsg1 is an essential GTPase predominantly located in the endoplasmic reticulum and, in some cells, in Cajal bodies in the nucleus. Comparison of localization and siRNA datasets suggests that all members of the family are essential GTPases that have increased in number as the compartmentalization of the eukaryotic cell and the ribosome biogenesis pathway have evolved.
Conclusion: We propose a scenario, consistent with our data, for the evolution of this family: cytoplasmic components were first acquired, followed by nuclear components, and finally the mitochondrial and chloroplast elements were derived from different bacterial species, in parallel with the formation of the nucleolus and the specialization of nuclear components.
Figures
Similar articles
-
Reading the Evolution of Compartmentalization in the Ribosome Assembly Toolbox: The YRG Protein Family.PLoS One. 2017 Jan 10;12(1):e0169750. doi: 10.1371/journal.pone.0169750. eCollection 2017. PLoS One. 2017. PMID: 28072865 Free PMC article.
-
Small GTPases and the evolution of the eukaryotic cell.Bioessays. 2003 Nov;25(11):1129-38. doi: 10.1002/bies.10353. Bioessays. 2003. PMID: 14579253
-
MitoRes: a resource of nuclear-encoded mitochondrial genes and their products in Metazoa.BMC Bioinformatics. 2006 Jan 24;7:36. doi: 10.1186/1471-2105-7-36. BMC Bioinformatics. 2006. PMID: 16433928 Free PMC article.
-
Biochemical analyses of the Wrch atypical Rho family GTPases.Methods Enzymol. 2006;406:11-26. doi: 10.1016/S0076-6879(06)06002-2. Methods Enzymol. 2006. PMID: 16472646 Review.
-
The function and diversity of plastid protein import pathways: a multilane GTPase highway into plastids.Traffic. 2006 Mar;7(3):248-57. doi: 10.1111/j.1600-0854.2005.00382.x. Traffic. 2006. PMID: 16497220 Review.
Cited by
-
The nucleolar protein GNL3 prevents resection of stalled replication forks.EMBO Rep. 2023 Dec 6;24(12):e57585. doi: 10.15252/embr.202357585. Epub 2023 Nov 15. EMBO Rep. 2023. PMID: 37965896 Free PMC article.
-
Nucleostemin: Another nucleolar "Twister" of the p53-MDM2 loop.Cell Cycle. 2010 Aug 15;9(16):3227-32. doi: 10.4161/cc.9.16.12605. Epub 2010 Aug 4. Cell Cycle. 2010. PMID: 20703089 Free PMC article.
-
Nmd3 is a structural mimic of eIF5A, and activates the cpGTPase Lsg1 during 60S ribosome biogenesis.EMBO J. 2017 Apr 3;36(7):854-868. doi: 10.15252/embj.201696012. Epub 2017 Feb 8. EMBO J. 2017. PMID: 28179369 Free PMC article.
-
Landscape of RNA-binding proteins in diagnostic utility, immune cell infiltration and PANoptosis features of heart failure.Front Genet. 2022 Oct 14;13:1004163. doi: 10.3389/fgene.2022.1004163. eCollection 2022. Front Genet. 2022. PMID: 36313471 Free PMC article.
-
Regulation of ribosome biogenesis by nucleostemin 3 promotes local and systemic growth in Drosophila.Genetics. 2013 May;194(1):101-15. doi: 10.1534/genetics.112.149104. Epub 2013 Feb 22. Genetics. 2013. PMID: 23436180 Free PMC article.
References
-
- Giaever G, Chu AM, Ni L, Connelly C, Riles L, Veronneau S, Dow S, Lucau-Danila A, Anderson K, Andre B, Arkin AP, Astromoff A, El-Bakkoury M, Bangham R, Benito R, Brachat S, Campanaro S, Curtiss M, Davis K, Deutschbauer A, Entian KD, Flaherty P, Foury F, Garfinkel DJ, Gerstein M, Gotte D, Guldener U, Hegemann JH, Hempel S, Herman Z, Jaramillo DF, Kelly DE, Kelly SL, Kotter P, LaBonte D, Lamb DC, Lan N, Liang H, Liao H, Liu L, Luo C, Lussier M, Mao R, Menard P, Ooi SL, Revuelta JL, Roberts CJ, Rose M, Ross-Macdonald P, Scherens B, Schimmack G, Shafer B, Shoemaker DD, Sookhai-Mahadeo S, Storms RK, Strathern JN, Valle G, Voet M, Volckaert G, Wang CY, Ward TR, Wilhelmy J, Winzeler EA, Yang Y, Yen G, Youngman E, Yu K, Bussey H, Boeke JD, Snyder M, Philippsen P, Davis RW, Johnston M. Functional profiling of the Saccharomyces cerevisiae genome. Nature. 2002;418:387–391. doi: 10.1038/nature00935. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

