Autophagy in cell death: an innocent convict?
- PMID: 16200202
- PMCID: PMC1236698
- DOI: 10.1172/JCI26390
Autophagy in cell death: an innocent convict?
Erratum in
- J Clin Invest. 2006 Dec;116(12):3293
Abstract
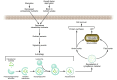
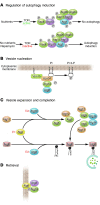
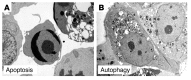
The visualization of autophagosomes in dying cells has led to the belief that autophagy is a nonapoptotic form of programmed cell death. This concept has now been evaluated using cells and organisms deficient in autophagy genes. Most evidence indicates that, at least in cells with intact apoptotic machinery, autophagy is primarily a pro-survival rather than a pro-death mechanism. This review summarizes the evidence linking autophagy to cell survival and cell death, the complex interplay between autophagy and apoptosis pathways, and the role of autophagy-dependent survival and death pathways in clinical diseases.
Figures
Similar articles
-
Autophagy and caspases: a new cell death program.Cell Cycle. 2004 Sep;3(9):1124-6. Epub 2004 Sep 20. Cell Cycle. 2004. PMID: 15326383 Review.
-
Crosstalk between autophagy and apoptosis in heart disease.Circ Res. 2008 Aug 15;103(4):343-51. doi: 10.1161/CIRCRESAHA.108.175448. Circ Res. 2008. PMID: 18703786 Review.
-
The complex interplay between autophagy and cell death pathways.Biochem J. 2022 Jan 14;479(1):75-90. doi: 10.1042/BCJ20210450. Biochem J. 2022. PMID: 35029627 Review.
-
Does autophagy contribute to cell death?Autophagy. 2005 Jul;1(2):66-74. doi: 10.4161/auto.1.2.1738. Epub 2005 Jul 13. Autophagy. 2005. PMID: 16874022 Review.
-
The role of cell signaling in the crosstalk between autophagy and apoptosis in the regulation of tumor cell survival in response to sorafenib and neratinib.Semin Cancer Biol. 2020 Nov;66:129-139. doi: 10.1016/j.semcancer.2019.10.013. Epub 2019 Oct 20. Semin Cancer Biol. 2020. PMID: 31644944 Free PMC article. Review.
Cited by
-
Cordycepin Activates Autophagy to Suppress FGF9-induced TM3 Mouse Leydig Progenitor Cell Proliferation.Cancer Genomics Proteomics. 2024 Nov-Dec;21(6):630-644. doi: 10.21873/cgp.20479. Cancer Genomics Proteomics. 2024. PMID: 39467624 Free PMC article.
-
Suppression of autophagy by CUB domain-containing protein 1 signaling is essential for anchorage-independent survival of lung cancer cells.Cancer Sci. 2013 Jul;104(7):865-70. doi: 10.1111/cas.12154. Epub 2013 Apr 19. Cancer Sci. 2013. PMID: 23510015 Free PMC article.
-
Ceramide induced mitophagy and tumor suppression.Biochim Biophys Acta. 2015 Oct;1853(10 Pt B):2834-45. doi: 10.1016/j.bbamcr.2014.12.039. Epub 2015 Jan 26. Biochim Biophys Acta. 2015. PMID: 25634657 Free PMC article. Review.
-
Cardiac aging: from molecular mechanisms to significance in human health and disease.Antioxid Redox Signal. 2012 Jun 15;16(12):1492-526. doi: 10.1089/ars.2011.4179. Epub 2012 Apr 3. Antioxid Redox Signal. 2012. PMID: 22229339 Free PMC article. Review.
-
MicroRNA-mediated autophagic signaling networks and cancer chemoresistance.Cancer Biother Radiopharm. 2013 Oct;28(8):573-8. doi: 10.1089/cbr.2012.1460. Epub 2013 Jul 10. Cancer Biother Radiopharm. 2013. PMID: 23841710 Free PMC article. Review.
References
-
- Levine B, Klionsky DJ. Development by self-digestion: molecular mechanisms and biological functions of autophagy. Dev. Cell. 2004;6:463–477. - PubMed
-
- Lum JJ, DeBarardinis RJ, Thompson CB. Autophagy in metazoans: cell survival in the land of plenty [review] Nat. Rev. Mol. Cell Biol. 2005;6:439–448. - PubMed
-
- Klionsky DJ, et al. A unified nomenclature for yeast autophagy-related genes. Dev. Cell. 2003;5:539–545. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

