Direct modulation of RNA polymerase core functions by basal transcription factors
- PMID: 16135821
- PMCID: PMC1234337
- DOI: 10.1128/MCB.25.18.8344-8355.2005
Direct modulation of RNA polymerase core functions by basal transcription factors
Abstract
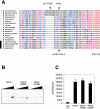
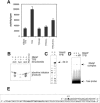
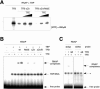
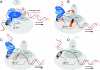
Archaeal RNA polymerases (RNAPs) are recruited to promoters through the joint action of three basal transcription factors: TATA-binding protein, TFB (archaeal homolog of TFIIB), and TFE (archaeal homolog of TFIIE). Our results demonstrate several new insights into the mechanisms of TFB and TFE during the transcription cycle. (i) The N-terminal Zn ribbon of TFB displays a surprising degree of redundancy for the recruitment of RNAP during transcription initiation in the archaeal system. (ii) The B-finger domain of TFB participates in transcription initiation events by stimulating abortive and productive transcription in a recruitment-independent function. TFB thus combines physical recruitment of the RNAP with an active role in influencing the catalytic properties of RNAP during transcription initiation. (iii) TFB mutations are complemented by TFE, thereby demonstrating that both factors act synergistically during transcription initiation. (iv) An additional function of TFE is to dynamically alter the nucleic acid-binding properties of RNAP by stabilizing the initiation complex and destabilizing elongation complexes.
Figures


Similar articles
-
Transcription factor B contacts promoter DNA near the transcription start site of the archaeal transcription initiation complex.J Biol Chem. 2004 Jan 23;279(4):2825-31. doi: 10.1074/jbc.M311433200. Epub 2003 Nov 3. J Biol Chem. 2004. PMID: 14597623
-
Modulation of RNA polymerase core functions by basal transcription factor TFB/TFIIB.Biochem Soc Symp. 2006;(73):49-58. doi: 10.1042/bss0730049. Biochem Soc Symp. 2006. PMID: 16626286
-
The linker domain of basal transcription factor TFIIB controls distinct recruitment and transcription stimulation functions.Nucleic Acids Res. 2011 Jan;39(2):464-74. doi: 10.1093/nar/gkq809. Epub 2010 Sep 17. Nucleic Acids Res. 2011. PMID: 20851833 Free PMC article.
-
Archaeal chromatin and transcription.Mol Microbiol. 2003 May;48(3):587-98. doi: 10.1046/j.1365-2958.2003.03439.x. Mol Microbiol. 2003. PMID: 12694606 Review.
-
Determinants of transcription initiation by archaeal RNA polymerase.Curr Opin Microbiol. 2005 Dec;8(6):677-84. doi: 10.1016/j.mib.2005.10.016. Epub 2005 Oct 24. Curr Opin Microbiol. 2005. PMID: 16249119 Review.
Cited by
-
Displacement of the transcription factor B reader domain during transcription initiation.Nucleic Acids Res. 2018 Nov 2;46(19):10066-10081. doi: 10.1093/nar/gky699. Nucleic Acids Res. 2018. PMID: 30102372 Free PMC article.
-
The initiation factor TFE and the elongation factor Spt4/5 compete for the RNAP clamp during transcription initiation and elongation.Mol Cell. 2011 Jul 22;43(2):263-74. doi: 10.1016/j.molcel.2011.05.030. Mol Cell. 2011. PMID: 21777815 Free PMC article.
-
TAF1B is a TFIIB-like component of the basal transcription machinery for RNA polymerase I.Science. 2011 Sep 16;333(6049):1640-2. doi: 10.1126/science.1207656. Science. 2011. PMID: 21921199 Free PMC article.
-
The transcript cleavage factor paralogue TFS4 is a potent RNA polymerase inhibitor.Nat Commun. 2017 Dec 4;8(1):1914. doi: 10.1038/s41467-017-02081-3. Nat Commun. 2017. PMID: 29203770 Free PMC article.
-
Cbp1 and Cren7 form chromatin-like structures that ensure efficient transcription of long CRISPR arrays.Nat Commun. 2024 Feb 22;15(1):1620. doi: 10.1038/s41467-024-45728-8. Nat Commun. 2024. PMID: 38388540 Free PMC article.
References
-
- Bartlett, M. S., M. Thomm, and E. P. Geiduschek. 2004. Topography of the euryarchaeal transcription initiation complex. J. Biol. Chem. 279:894-903. - PubMed
-
- Bell, S. D., and S. P. Jackson. 2000. The role of transcription factor B in transcription initiation and promoter clearance in the archaeon Sulfolobus acidocaldarius. J. Biol. Chem. 275:12934-12940. - PubMed
-
- Bell, S. D., and S. P. Jackson. 2001. Mechanism and regulation of transcription in archaea. Curr. Opin. Microbiol. 4:208-213. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
