Regulation of NuA4 histone acetyltransferase activity in transcription and DNA repair by phosphorylation of histone H4
- PMID: 16135807
- PMCID: PMC1234332
- DOI: 10.1128/MCB.25.18.8179-8190.2005
Regulation of NuA4 histone acetyltransferase activity in transcription and DNA repair by phosphorylation of histone H4
Abstract
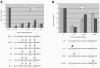
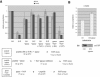
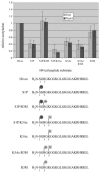
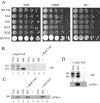
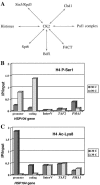
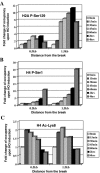
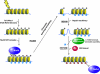
The NuA4 complex is a histone H4/H2A acetyltransferase involved in transcription and DNA repair. While histone acetylation is important in many processes, it has become increasingly clear that additional histone modifications also play a crucial interrelated role. To understand how NuA4 action is regulated, we tested various H4 tail peptides harboring known modifications in HAT assays. While dimethylation at arginine 3 (R3M) had little effect on NuA4 activity, phosphorylation of serine 1 (S1P) strongly decreased the ability of the complex to acetylate H4 peptides. However, R3M in combination with S1P alleviates the repression of NuA4 activity. Chromatin from cells treated with DNA damage-inducing agents shows an increase in phosphorylation of serine 1 and a concomitant decrease in H4 acetylation. We found that casein kinase 2 phosphorylates histone H4 and associates with the Rpd3 deacetylase complex, demonstrating a physical connection between phosphorylation of serine 1 and unacetylated H4 tails. Chromatin immunoprecipitation experiments also link local phosphorylation of H4 with its deacetylation, during both transcription and DNA repair. Time course chromatin immunoprecipitation data support a model in which histone H4 phosphorylation occurs after NuA4 action during double-strand break repair at the step of chromatin restoration and deacetylation. These findings demonstrate that H4 phospho-serine 1 regulates chromatin acetylation by the NuA4 complex and that this process is important for normal gene expression and DNA repair.
Figures

Similar articles
-
Role of an ING1 growth regulator in transcriptional activation and targeted histone acetylation by the NuA4 complex.Mol Cell Biol. 2001 Nov;21(22):7629-40. doi: 10.1128/MCB.21.22.7629-7640.2001. Mol Cell Biol. 2001. PMID: 11604499 Free PMC article.
-
Acetylation of histone H4 by Esa1 is required for DNA double-strand break repair.Nature. 2002 Sep 26;419(6905):411-5. doi: 10.1038/nature01035. Nature. 2002. PMID: 12353039
-
NuA4, an essential transcription adaptor/histone H4 acetyltransferase complex containing Esa1p and the ATM-related cofactor Tra1p.EMBO J. 1999 Sep 15;18(18):5108-19. doi: 10.1093/emboj/18.18.5108. EMBO J. 1999. PMID: 10487762 Free PMC article.
-
Roles of histone acetyltransferases and deacetylases in gene regulation.Bioessays. 1998 Aug;20(8):615-26. doi: 10.1002/(SICI)1521-1878(199808)20:8<615::AID-BIES4>3.0.CO;2-H. Bioessays. 1998. PMID: 9780836 Review.
-
Histone acetylation and deacetylation in yeast.Nat Rev Mol Cell Biol. 2003 Apr;4(4):276-84. doi: 10.1038/nrm1075. Nat Rev Mol Cell Biol. 2003. PMID: 12671650 Review.
Cited by
-
Catalysis-dependent and redundant roles of Dma1 and Dma2 in maintenance of genome stability in Saccharomyces cerevisiae.J Biol Chem. 2021 Jan-Jun;296:100721. doi: 10.1016/j.jbc.2021.100721. Epub 2021 Apr 29. J Biol Chem. 2021. PMID: 33933452 Free PMC article.
-
Probing nucleosome function: a highly versatile library of synthetic histone H3 and H4 mutants.Cell. 2008 Sep 19;134(6):1066-78. doi: 10.1016/j.cell.2008.07.019. Cell. 2008. PMID: 18805098 Free PMC article.
-
Single-Molecule Investigations on Histone H2A-H2B Dynamics in the Nucleosome.Biochemistry. 2017 Feb 21;56(7):977-985. doi: 10.1021/acs.biochem.6b01252. Epub 2017 Feb 8. Biochemistry. 2017. PMID: 28128545 Free PMC article.
-
NatD promotes lung cancer progression by preventing histone H4 serine phosphorylation to activate Slug expression.Nat Commun. 2017 Oct 13;8(1):928. doi: 10.1038/s41467-017-00988-5. Nat Commun. 2017. PMID: 29030587 Free PMC article.
-
ATM-mediated transcriptional and developmental responses to gamma-rays in Arabidopsis.PLoS One. 2007 May 9;2(5):e430. doi: 10.1371/journal.pone.0000430. PLoS One. 2007. PMID: 17487278 Free PMC article.
References
-
- An, W., J. Kim, and R. G. Roeder. 2004. Ordered cooperative functions of PRMT1, p300, and CARM1 in transcriptional activation by p53. Cell 117:735-748. - PubMed
-
- Barber, C. M., F. B. Turner, Y. Wang, K. Hagstrom, S. D. Taverna, S. Mollah, B. Ueberheide, B. J. Meyer, D. F. Hunt, P. Cheung, and C. D. Allis. 2004. The enhancement of histone H4 and H2A serine 1 phosphorylation during mitosis and S-phase is evolutionarily conserved. Chromosoma 112:360-371. - PubMed
-
- Bird, A. W., D. Y. Yu, M. G. Pray-Grant, Q. Qiu, K. E. Harmon, P. C. Megee, P. A. Grant, M. M. Smith, and M. F. Christman. 2002. Acetylation of histone H4 by Esa1 is required for DNA double-strand break repair. Nature 419:411-415. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
