p53 isoforms can regulate p53 transcriptional activity
- PMID: 16131611
- PMCID: PMC1221884
- DOI: 10.1101/gad.1339905
p53 isoforms can regulate p53 transcriptional activity
Abstract
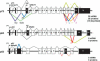
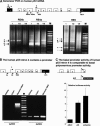
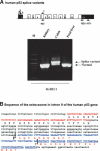
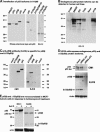
The recently discovered p53-related genes, p73 and p63, express multiple splice variants and N-terminally truncated forms initiated from an alternative promoter in intron 3. To date, no alternative promoter and multiple splice variants have been described for the p53 gene. In this study, we show that p53 has a gene structure similar to the p73 and p63 genes. The human p53 gene contains an alternative promoter and transcribes multiple splice variants. We show that p53 variants are expressed in normal human tissue in a tissue-dependent manner. We determine that the alternative promoter is conserved through evolution from Drosophila to man, suggesting that the p53 family gene structure plays an essential role in the multiple activities of the p53 family members. Consistent with this hypothesis, p53 variants are differentially expressed in human breast tumors compared with normal breast tissue. We establish that p53beta can bind differentially to promoters and can enhance p53 target gene expression in a promoter-dependent manner, while Delta133p53 is dominant-negative toward full-length p53, inhibiting p53-mediated apoptosis. The differential expression of the p53 isoforms in human tumors may explain the difficulties in linking p53 status to the biological properties and drug sensitivity of human cancer.
Figures



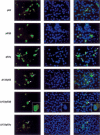
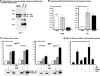
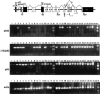
Comment in
-
p53: link to the past, bridge to the future.Genes Dev. 2005 Sep 15;19(18):2091-9. doi: 10.1101/gad.1362905. Genes Dev. 2005. PMID: 16166374 Review. No abstract available.
-
The continuing saga of p53--more sleepless nights ahead.Mol Cell. 2005 Sep 16;19(6):719-21. doi: 10.1016/j.molcel.2005.08.024. Mol Cell. 2005. PMID: 16168367
Similar articles
-
p53: link to the past, bridge to the future.Genes Dev. 2005 Sep 15;19(18):2091-9. doi: 10.1101/gad.1362905. Genes Dev. 2005. PMID: 16166374 Review. No abstract available.
-
The neurogene BTG2TIS21/PC3 is transactivated by DeltaNp73alpha via p53 specifically in neuroblastoma cells.J Cell Sci. 2005 Mar 15;118(Pt 6):1245-53. doi: 10.1242/jcs.01704. Epub 2005 Mar 1. J Cell Sci. 2005. PMID: 15741235
-
Distinct expression of Survivin splice variants in breast carcinomas.Int J Oncol. 2005 Oct;27(4):1151-7. Int J Oncol. 2005. PMID: 16142334
-
Negative regulation of chemokine receptor CXCR4 by tumor suppressor p53 in breast cancer cells: implications of p53 mutation or isoform expression on breast cancer cell invasion.Oncogene. 2007 May 17;26(23):3329-37. doi: 10.1038/sj.onc.1210120. Epub 2006 Nov 27. Oncogene. 2007. PMID: 17130833
-
The isoforms of the p53 protein.Cold Spring Harb Perspect Biol. 2010 Mar;2(3):a000927. doi: 10.1101/cshperspect.a000927. Cold Spring Harb Perspect Biol. 2010. PMID: 20300206 Free PMC article. Review.
Cited by
-
Navigating the complexity of p53-DNA binding: implications for cancer therapy.Biophys Rev. 2024 Jul 11;16(4):479-496. doi: 10.1007/s12551-024-01207-4. eCollection 2024 Aug. Biophys Rev. 2024. PMID: 39309126 Free PMC article. Review.
-
Δ113p53/Δ133p53: survival and integrity.Oncotarget. 2015 Jun 10;6(16):13850-1. doi: 10.18632/oncotarget.4371. Oncotarget. 2015. PMID: 26116838 Free PMC article. No abstract available.
-
P53 aggregation, interactions with tau, and impaired DNA damage response in Alzheimer's disease.Acta Neuropathol Commun. 2020 Aug 10;8(1):132. doi: 10.1186/s40478-020-01012-6. Acta Neuropathol Commun. 2020. PMID: 32778161 Free PMC article.
-
Prognostic interaction between expression of p53 and estrogen receptor in patients with node-negative breast cancer: results from IBCSG Trials VIII and IX.Breast Cancer Res. 2012 Nov 5;14(6):R143. doi: 10.1186/bcr3348. Breast Cancer Res. 2012. PMID: 23127292 Free PMC article. Clinical Trial.
-
Comparison of Three Different Methods for Determining Cell Proliferation in Breast Cancer Cell Lines.J Vis Exp. 2016 Sep 3;(115):54350. doi: 10.3791/54350. J Vis Exp. 2016. PMID: 27685661 Free PMC article.
References
-
- Almog N., Goldfinger, N., and Rotter, V. 2000. p53-dependent apoptosis is regulated by a C-terminally alternatively spliced form of murine p53. Oncogene 19: 3395-3403. - PubMed
-
- Bates G.J., Nicol, S.M., Wilson, B.J., Jacobs, A.M., Bourdon, J.C., Wardrop, J., Gregory, D.J., Lane, D.P., Perkins, N.D., and Fuller-Pace, F.V. 2005. The DEAD box protein p68: A novel transcriptional coactivator of the p53 tumour suppressor. EMBO J. 24: 543-553. [Epub 2005 January 20.] - PMC - PubMed
-
- Benard J., Douc-Rasy, S., and Ahomadegbe, J.C. 2003. TP53 family members and human cancers. Hum. Mutat. 21: 182-191. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous
