Competitive hybridization kinetics reveals unexpected behavior patterns
- PMID: 16126833
- PMCID: PMC1366793
- DOI: 10.1529/biophysj.104.058552
Competitive hybridization kinetics reveals unexpected behavior patterns
Abstract
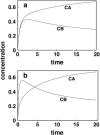
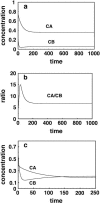
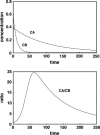
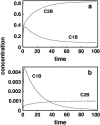
Although the kinetics of hybridization between a soluble polynucleotide and an immobilized complementary sequence have been studied by others, it is almost universally assumed that the interaction between each probe/target pair can be treated as a separate event. This simplifies the mathematics considerably, but it can give a false picture of the extent of hybridization that one achieves at equilibrium as well as the relative quantities of each hybridized pair during the approach to equilibrium. Here we solve the relevant kinetics equations simultaneously using Mathematica as a simulation language. Among the interesting results of this study are that, for certain circumstances, the relative ratio of incorrect to correct hybrids can change dramatically with time; that the relative abundances of two pairs are not what one would expect based on their equilibrium dissociation constants; that the volume of a wash solution after hybridization can have a large effect on results; and the fact that a short wash is typically better than a long one. We show that an optimum wash time exists for a given set of conditions. In addition, the ratio of soluble to insoluble (spotted) molecules can influence results substantially. Finally, the true levels of rare transcripts can be masked by the presence of highly abundant ones. Code is supplied to enable others to study conditions beyond those presented in this article.
Figures
Similar articles
-
Competitive displacement of DNA during surface hybridization.Biophys J. 2007 Jan 1;92(1):L10-2. doi: 10.1529/biophysj.106.097121. Epub 2006 Oct 20. Biophys J. 2007. PMID: 17056736 Free PMC article.
-
Control of complexity constraints on combinatorial screening for preferred oligonucleotide hybridization sites on structured RNA.Biochemistry. 1997 Apr 22;36(16):5004-19. doi: 10.1021/bi9620767. Biochemistry. 1997. PMID: 9125523
-
The biophysics of DNA hybridization with immobilized oligonucleotide probes.Biophys J. 1995 Dec;69(6):2243-55. doi: 10.1016/S0006-3495(95)80095-0. Biophys J. 1995. PMID: 8599632 Free PMC article.
-
DNA probes: applications of the principles of nucleic acid hybridization.Crit Rev Biochem Mol Biol. 1991;26(3-4):227-59. doi: 10.3109/10409239109114069. Crit Rev Biochem Mol Biol. 1991. PMID: 1718662 Review.
-
Mismatch formation in solution and on DNA microarrays: how modified nucleosides can overcome shortcomings of imperfect hybridization caused by oligonucleotide composition and base pairing.Mol Biosyst. 2008 Mar;4(3):232-45. doi: 10.1039/b713259j. Epub 2008 Feb 4. Mol Biosyst. 2008. PMID: 18437266 Review.
Cited by
-
An efficient algorithm for the stochastic simulation of the hybridization of DNA to microarrays.BMC Bioinformatics. 2009 Dec 10;10:411. doi: 10.1186/1471-2105-10-411. BMC Bioinformatics. 2009. PMID: 20003312 Free PMC article.
-
Kinetics of multiplex hybridization: mechanisms and implications.Biophys J. 2008 Mar 1;94(5):1726-34. doi: 10.1529/biophysj.107.121459. Epub 2007 Nov 9. Biophys J. 2008. PMID: 17993490 Free PMC article.
-
Competitive displacement of DNA during surface hybridization.Biophys J. 2007 Jan 1;92(1):L10-2. doi: 10.1529/biophysj.106.097121. Epub 2006 Oct 20. Biophys J. 2007. PMID: 17056736 Free PMC article.
-
A competitive kinetic model of nucleic acid surface hybridization in the presence of point mutants.Biophys J. 2006 Feb 1;90(3):831-40. doi: 10.1529/biophysj.105.072314. Epub 2005 Nov 11. Biophys J. 2006. PMID: 16284267 Free PMC article.
-
Microarray analysis of cytoplasmic versus whole cell RNA reveals a considerable number of missed and false positive mRNAs.RNA. 2009 Oct;15(10):1917-28. doi: 10.1261/rna.1677409. Epub 2009 Aug 24. RNA. 2009. PMID: 19703940 Free PMC article.
References
-
- Dorris, D. R., A. Nguyen, L. Gieser, R. Lockner, A. Lublinsky, M. Patterson, E. Touma, T. J. Sendera, R. Elghanian, and A. Mazumder. 2003. Oligodeoxyribonucleotide probe accessibility on a three-dimensional DNA microarray surface and the effect of hybridization time on the accuracy of expression ratios. BMC Biotechnol. 3:6. - PMC - PubMed
-
- Lauffenburger, D. A., and J. J. Linderman. 1993. Receptors. Oxford University Press, Oxford, UK.
-
- Graves, D. J. 1999. Microarrays: powerful tools for genetic analysis come of age. Trends Biotechnol. 17:127–134. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

