Sequence conservation, relative isoform frequencies, and nonsense-mediated decay in evolutionarily conserved alternative splicing
- PMID: 16123126
- PMCID: PMC1192826
- DOI: 10.1073/pnas.0506139102
Sequence conservation, relative isoform frequencies, and nonsense-mediated decay in evolutionarily conserved alternative splicing
Abstract
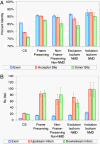
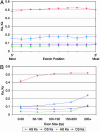
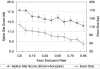
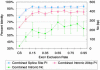
Studies of expressed sequence tag data sets have revealed large numbers of splicing variants for human genes, but it remains challenging to distinguish functionally important variants from aberrant splicing, clarify the nature of the alternative functions, and understand the signals that regulate splicing choices. To help address these issues, we have constructed and analyzed a large data set of 1,478 exon-skipping alternative splicing (AS) variants evolutionarily conserved in human and mouse. In about one-fifth of cases, one isoform appears subject to nonsense-mediated mRNA decay (NMD), supporting the idea that a major role of AS is to regulate gene expression; one-quarter of these NMD-inducing cases involve a conserved exon whose apparent sole purpose is to mediate destruction of the message when included. We explore sequence conservation likely related to splicing regulation, using in part a measure of the overall amount of conserved information in a sequence, and find that the increased conservation that has been observed within AS exons primarily affects synonymous sites, suggesting that regulatory signals significantly constrain synonymous substitution rates. We show that a lower frequency of the inclusion isoform relative to the exclusion isoform tends to be associated with weaker splice site signals, smaller exon size, and higher intronic sequence conservation, and provide evidence that all of these factors are under selection to control relative isoform frequencies. Some conserved instances of AS appear to represent aberrant splicing events that by chance have occurred in both species, and we develop a nonparametric likelihood approach to identify these.
Figures

Similar articles
-
Alternative splicing and nonsense-mediated mRNA decay in the regulation of a new adenomatous polyposis coli transcript.Gene. 2007 Jun 15;395(1-2):8-14. doi: 10.1016/j.gene.2006.10.027. Epub 2007 Jan 12. Gene. 2007. PMID: 17360132
-
Unusual intron conservation near tissue-regulated exons found by splicing microarrays.PLoS Comput Biol. 2006 Jan;2(1):e4. doi: 10.1371/journal.pcbi.0020004. Epub 2006 Jan 20. PLoS Comput Biol. 2006. PMID: 16424921 Free PMC article.
-
A minor alternative transcript of the fumarylacetoacetate hydrolase gene produces a protein despite being likely subjected to nonsense-mediated mRNA decay.BMC Mol Biol. 2005 Jan 7;6:1. doi: 10.1186/1471-2199-6-1. BMC Mol Biol. 2005. PMID: 15638932 Free PMC article.
-
Alternative splicing of mutually exclusive exons--a review.Biosystems. 2013 Oct;114(1):31-8. doi: 10.1016/j.biosystems.2013.07.003. Epub 2013 Jul 11. Biosystems. 2013. PMID: 23850531 Review.
-
How prevalent is functional alternative splicing in the human genome?Trends Genet. 2004 Feb;20(2):68-71. doi: 10.1016/j.tig.2003.12.004. Trends Genet. 2004. PMID: 14746986 Review.
Cited by
-
The endogenous HBZ interactome in ATL leukemic cells reveals an unprecedented complexity of host interacting partners involved in RNA splicing.Front Immunol. 2022 Aug 1;13:939863. doi: 10.3389/fimmu.2022.939863. eCollection 2022. Front Immunol. 2022. PMID: 35979358 Free PMC article.
-
Origin of spliceosomal introns and alternative splicing.Cold Spring Harb Perspect Biol. 2014 Jun 2;6(6):a016071. doi: 10.1101/cshperspect.a016071. Cold Spring Harb Perspect Biol. 2014. PMID: 24890509 Free PMC article.
-
Highly variable mRNA expression and splicing of L-type voltage-dependent calcium channel alpha subunit 1C in human heart tissues.Pharmacogenet Genomics. 2006 Oct;16(10):735-45. doi: 10.1097/01.fpc.0000230119.34205.8a. Pharmacogenet Genomics. 2006. PMID: 17001293 Free PMC article.
-
A novel tissue-specific alternatively spliced form of the A-to-I RNA editing enzyme ADAR2.RNA Biol. 2010 Mar-Apr;7(2):253-62. doi: 10.4161/rna.7.2.11568. Epub 2010 Mar 17. RNA Biol. 2010. PMID: 20215858 Free PMC article.
-
Evolutionary conservation and regulation of particular alternative splicing events in plant SR proteins.Nucleic Acids Res. 2006;34(16):4395-405. doi: 10.1093/nar/gkl570. Epub 2006 Aug 26. Nucleic Acids Res. 2006. PMID: 16936312 Free PMC article.
References
-
- Graveley, B. R. (2001) Trends Genet. 17, 100–107. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

