Kinetic resolution of bimolecular hybridization versus intramolecular folding in nucleic acids by surface plasmon resonance: application to G-quadruplex/duplex competition in human c-myc promoter
- PMID: 16085756
- PMCID: PMC1183106
- DOI: 10.1093/nar/gki750
Kinetic resolution of bimolecular hybridization versus intramolecular folding in nucleic acids by surface plasmon resonance: application to G-quadruplex/duplex competition in human c-myc promoter
Abstract
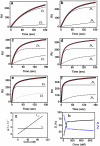
The human oncogene c-myc is regulated by G-quadruplex formation within the nuclease hypersensitive element (NHE III(I)) in the c-myc promoter, making the quadruplex a strong anti-cancer target. With respect to this, the competing equilibrium between intramolecular quadruplex folding and bimolecular duplex formation is poorly understood and very few techniques have addressed this problem. We present a method for simultaneously determining the kinetic constants for G-quadruplex folding/unfolding and hybridization in the presence of the complementary strand from a single reaction using an optical biosensor based on surface plasmon resonance (SPR). Using this technique, we demonstrate for the first time that quadruplex formation in the c-myc promoter is favored at low strand concentrations. Our results indicate favorable quadruplex folding (equilibrium folding constant K(F) of 2.09 calculated from the kinetic parameters: folding rate constant, k(f) = 1.65 x 10(-2) s(-1) and unfolding rate constant, k(u) = 7.90 x 10(-3) s(-1)) in 150 mM K+. The hybridization rate constants detected concurrently gave a bimolecular association constant, k(a) = 1.37 x 10(5) M(-1) s(-1) and dissociation constant, k(d) = 4.94 x 10(-5) s(-1). Interestingly, in the presence of Na+ we observed that G-quadruplex folding was unfavorable (K(F) = 0.54). Implication of our results on the c-myc transcription activation model is discussed in light of aberrant c-myc expression observed on destabilization of the G-quadruplex.
Figures

Similar articles
-
Determining the folding and unfolding rate constants of nucleic acids by biosensor. Application to telomere G-quadruplex.J Am Chem Soc. 2004 Oct 20;126(41):13255-64. doi: 10.1021/ja048398c. J Am Chem Soc. 2004. PMID: 15479079
-
Quadruplex-duplex competition in the nuclease hypersensitive element of human c-myc promoter: C to T mutation in C-rich strand enhances duplex association.Biochem Biophys Res Commun. 2005 Feb 4;327(1):49-56. doi: 10.1016/j.bbrc.2004.11.137. Biochem Biophys Res Commun. 2005. PMID: 15629428
-
Kinetic and thermodynamic characterization of telomeric G-quadruplex by nonequilibrium capillary electrophoresis: application to G-quadruplex/duplex competition.Anal Chem. 2008 Sep 15;80(18):6935-41. doi: 10.1021/ac801335y. Epub 2008 Aug 12. Anal Chem. 2008. PMID: 18693771
-
Drug targeting of the c-MYC promoter to repress gene expression via a G-quadruplex silencer element.Semin Oncol. 2006 Aug;33(4):498-512. doi: 10.1053/j.seminoncol.2006.04.012. Semin Oncol. 2006. PMID: 16890804 Review.
-
Thermodynamic and kinetic characterization of the dissociation and assembly of quadruplex nucleic acids.Biopolymers. 2000-2001;56(3):147-94. doi: 10.1002/1097-0282(2000/2001)56:3<147::AID-BIP10011>3.0.CO;2-N. Biopolymers. 2000. PMID: 11745110 Review.
Cited by
-
A recombination hot spot in HIV-1 contains guanosine runs that can form a G-quartet structure and promote strand transfer in vitro.J Biol Chem. 2009 Dec 4;284(49):33883-93. doi: 10.1074/jbc.M109.055368. Epub 2009 Oct 12. J Biol Chem. 2009. PMID: 19822521 Free PMC article.
-
DNA architecture: from G to Z.Curr Opin Struct Biol. 2006 Jun;16(3):288-98. doi: 10.1016/j.sbi.2006.05.011. Epub 2006 May 22. Curr Opin Struct Biol. 2006. PMID: 16714104 Free PMC article. Review.
-
Genome-wide analyses of recombination prone regions predict role of DNA structural motif in recombination.PLoS One. 2009;4(2):e4399. doi: 10.1371/journal.pone.0004399. Epub 2009 Feb 9. PLoS One. 2009. PMID: 19198658 Free PMC article.
-
A parallel quadruplex DNA is bound tightly but unfolded slowly by pif1 helicase.J Biol Chem. 2015 Mar 6;290(10):6482-94. doi: 10.1074/jbc.M114.630749. Epub 2015 Jan 14. J Biol Chem. 2015. PMID: 25589786 Free PMC article.
-
QGRS Mapper: a web-based server for predicting G-quadruplexes in nucleotide sequences.Nucleic Acids Res. 2006 Jul 1;34(Web Server issue):W676-82. doi: 10.1093/nar/gkl253. Nucleic Acids Res. 2006. PMID: 16845096 Free PMC article.
References
-
- Pelengaris S., Rudolph B., Littlewood T. Action of Myc in vivo—proliferation and apoptosis. Curr. Opin. Genet. Dev. 2000;10:100–105. - PubMed
-
- Spencer C.A., Groudine M. Control of c-myc regulation in normal and neoplastic cells. Adv. Cancer Res. 1991;56:1–48. - PubMed
-
- Facchini L.M., Penn L.Z. The molecular role of Myc in growth and transformation: recent discoveries lead to new insights. FASEB J. 1998;12:633–651. - PubMed
-
- Marcu K.B., Bossone S.A., Patel A.J. myc function and regulation. Annu. Rev. Biochem. 1992;61:809–860. - PubMed

