Structure of a human autoimmune TCR bound to a myelin basic protein self-peptide and a multiple sclerosis-associated MHC class II molecule
- PMID: 16079912
- PMCID: PMC1201352
- DOI: 10.1038/sj.emboj.7600771
Structure of a human autoimmune TCR bound to a myelin basic protein self-peptide and a multiple sclerosis-associated MHC class II molecule
Abstract
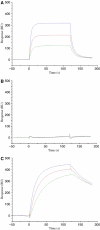
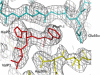
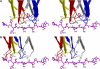
Multiple sclerosis is mediated by T-cell responses to central nervous system antigens such as myelin basic protein (MBP). To investigate self-peptide/major histocompatibility complex (MHC) recognition and T-cell receptor (TCR) degeneracy, we determined the crystal structure, at 2.8 A resolution, of an autoimmune TCR (3A6) bound to an MBP self-peptide and the multiple sclerosis-associated MHC class II molecule, human leukocyte antigen (HLA)-DR2a. The complex reveals that 3A6 primarily recognizes the N-terminal portion of MBP, in contrast with antimicrobial and alloreactive TCRs, which focus on the peptide center. Moreover, this binding mode, which may be frequent among autoimmune TCRs, is compatible with a wide range of orientation angles of TCR to peptide/MHC. The interface is characterized by a scarcity of hydrogen bonds between TCR and peptide, and TCR-induced conformational changes in MBP/HLA-DR2a, which likely explain the low observed affinity. Degeneracy of 3A6, manifested by recognition of superagonist peptides bearing substitutions at nearly all TCR-contacting positions, results from the few specific interactions between 3A6 and MBP, allowing optimization of interface complementarity through variations in the peptide.
Figures
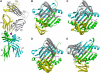

Similar articles
-
The 3A6-TCR/superagonist/HLA-DR2a complex shows similar interface and reduced flexibility compared to the complex with self-peptide.Proteins. 2020 Jan;88(1):31-46. doi: 10.1002/prot.25764. Epub 2019 Jun 25. Proteins. 2020. PMID: 31237711
-
Positioning of autoimmune TCR-Ob.2F3 and TCR-Ob.3D1 on the MBP85-99/HLA-DR2 complex.Proc Natl Acad Sci U S A. 2008 Oct 7;105(40):15523-8. doi: 10.1073/pnas.0807338105. Epub 2008 Sep 29. Proc Natl Acad Sci U S A. 2008. PMID: 18824684 Free PMC article.
-
Structural basis for the binding of an immunodominant peptide from myelin basic protein in different registers by two HLA-DR2 proteins.J Mol Biol. 2000 Nov 24;304(2):177-88. doi: 10.1006/jmbi.2000.4198. J Mol Biol. 2000. PMID: 11080454
-
The structural interactions between T cell receptors and MHC-peptide complexes place physical limits on self-nonself discrimination.Curr Top Microbiol Immunol. 2005;296:19-37. doi: 10.1007/3-540-30791-5_2. Curr Top Microbiol Immunol. 2005. PMID: 16329190 Review.
-
Structural requirements for binding of myelin basic protein (MBP) peptides to MHC II: effects on immune regulation.Curr Med Chem. 2005;12(13):1521-35. doi: 10.2174/0929867054039053. Curr Med Chem. 2005. PMID: 15974985 Review.
Cited by
-
TCR Signaling Emerges from the Sum of Many Parts.Front Immunol. 2012 Jun 25;3:159. doi: 10.3389/fimmu.2012.00159. eCollection 2012. Front Immunol. 2012. PMID: 22737151 Free PMC article.
-
Current and future immunomodulation strategies to restore tolerance in autoimmune diseases.Cold Spring Harb Perspect Biol. 2012 Nov 1;4(11):a007542. doi: 10.1101/cshperspect.a007542. Cold Spring Harb Perspect Biol. 2012. PMID: 23125012 Free PMC article. Review.
-
A novel loop domain in superantigens extends their T cell receptor recognition site.J Mol Biol. 2007 Aug 3;371(1):210-21. doi: 10.1016/j.jmb.2007.05.038. Epub 2007 May 18. J Mol Biol. 2007. PMID: 17560605 Free PMC article.
-
Protective Allele for Multiple Sclerosis HLA-DRB1*01:01 Provides Kinetic Discrimination of Myelin and Exogenous Antigenic Peptides.Front Immunol. 2020 Jan 17;10:3088. doi: 10.3389/fimmu.2019.03088. eCollection 2019. Front Immunol. 2020. PMID: 32010139 Free PMC article.
-
Flexible Players within the Sheaths: The Intrinsically Disordered Proteins of Myelin in Health and Disease.Cells. 2020 Feb 18;9(2):470. doi: 10.3390/cells9020470. Cells. 2020. PMID: 32085570 Free PMC article. Review.
References
-
- Alam SM, Travers PJ, Wung JL, Nasholds W, Redpath S, Jameson SC, Gascoigne NR (1996) T-cell receptor affinity and thymocyte positive selection. Nature 381: 616–620 - PubMed
-
- Bielekova B, Goodwin B, Richert N, Cortese I, Kondo T, Afshar G, Gran B, Eaton J, Antel J, Frank JA, McFarland HF, Martin R (2000) Encephalitogenic potential of myelin basic protein peptide (83–99) in multiple sclerosis: results of a phase II clinical trial with an altered peptide ligand. Nat Med 6: 1167–1175 - PubMed
-
- Brünger AT, Adams PD, Clore GM, DeLano WL, Gros P, Gross-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, Read RJ, Rice LM, Simonson T, Warren GL (1998) Crystallography and NMR systems: a new software suite for macromolecular structure determination. Acta Crystallogr D 54: 905–921 - PubMed
-
- Cull MG, Schatz PJ (2000) Biotinylation of proteins in vivo and in vitro using small peptide tags. Methods Enzymol 326: 430–440 - PubMed
-
- Davis MM, Boniface JJ, Reich Z, Lyons D, Hampl J, Arden B, Chien Y (1998) Ligand recognition by αβ T cell receptors. Annu Rev Immunol 16: 523–544 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

