Genome sequencing in microfabricated high-density picolitre reactors
- PMID: 16056220
- PMCID: PMC1464427
- DOI: 10.1038/nature03959
Genome sequencing in microfabricated high-density picolitre reactors
Erratum in
- Nature. 2006 May 4;441(7089):120. Ho, Chun He [corrected to Ho, Chun Heen]
Abstract
The proliferation of large-scale DNA-sequencing projects in recent years has driven a search for alternative methods to reduce time and cost. Here we describe a scalable, highly parallel sequencing system with raw throughput significantly greater than that of state-of-the-art capillary electrophoresis instruments. The apparatus uses a novel fibre-optic slide of individual wells and is able to sequence 25 million bases, at 99% or better accuracy, in one four-hour run. To achieve an approximately 100-fold increase in throughput over current Sanger sequencing technology, we have developed an emulsion method for DNA amplification and an instrument for sequencing by synthesis using a pyrosequencing protocol optimized for solid support and picolitre-scale volumes. Here we show the utility, throughput, accuracy and robustness of this system by shotgun sequencing and de novo assembly of the Mycoplasma genitalium genome with 96% coverage at 99.96% accuracy in one run of the machine.
Figures


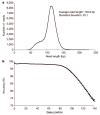
Comment in
-
Genomics: massively parallel sequencing.Nature. 2005 Sep 15;437(7057):326-7. doi: 10.1038/437326a. Nature. 2005. PMID: 16163333 No abstract available.
-
Massively parallel ("next-generation") DNA sequencing.Clin Chem. 2015 Jul;61(7):997-8. doi: 10.1373/clinchem.2014.237461. Epub 2015 Apr 28. Clin Chem. 2015. PMID: 25922443 Review. No abstract available.
Similar articles
-
Genomics: massively parallel sequencing.Nature. 2005 Sep 15;437(7057):326-7. doi: 10.1038/437326a. Nature. 2005. PMID: 16163333 No abstract available.
-
Massively parallel ("next-generation") DNA sequencing.Clin Chem. 2015 Jul;61(7):997-8. doi: 10.1373/clinchem.2014.237461. Epub 2015 Apr 28. Clin Chem. 2015. PMID: 25922443 Review. No abstract available.
-
The complete genome of an individual by massively parallel DNA sequencing.Nature. 2008 Apr 17;452(7189):872-6. doi: 10.1038/nature06884. Nature. 2008. PMID: 18421352
-
A method for parallel, automated, thermal cycling of submicroliter samples.Genome Res. 2001 Mar;11(3):441-7. doi: 10.1101/gr.gr1644r. Genome Res. 2001. PMID: 11230168 Free PMC article.
-
The use of capillary electrophoresis for DNA polymorphism analysis.Mol Biotechnol. 2003 May;24(1):41-68. doi: 10.1385/MB:24:1:41. Mol Biotechnol. 2003. PMID: 12721495 Review.
Cited by
-
Ancient orphan crop joins modern era: gene-based SNP discovery and mapping in lentil.BMC Genomics. 2013 Mar 18;14:192. doi: 10.1186/1471-2164-14-192. BMC Genomics. 2013. PMID: 23506258 Free PMC article.
-
Multifactorial diversity sustains microbial community stability.ISME J. 2013 Nov;7(11):2126-36. doi: 10.1038/ismej.2013.108. Epub 2013 Jul 4. ISME J. 2013. PMID: 23823494 Free PMC article.
-
Pharmacogenomics in children: advantages and challenges of next generation sequencing applications.Int J Pediatr. 2013;2013:136524. doi: 10.1155/2013/136524. Epub 2013 Jan 17. Int J Pediatr. 2013. PMID: 23401694 Free PMC article.
-
PvP01-DB: computational structural and functional characterization of soluble proteome of PvP01 strain of Plasmodium vivax.Database (Oxford). 2020 Jan 1;2020:baaa036. doi: 10.1093/database/baaa036. Database (Oxford). 2020. PMID: 32542363 Free PMC article.
-
Cancer treatment comes to age: from one-size-fits-all to next-generation sequencing (NGS) technologies.Bioimpacts. 2024;14(4):29957. doi: 10.34172/bi.2023.29957. Epub 2023 Dec 23. Bioimpacts. 2024. PMID: 39104623 Free PMC article. Review.
References
-
- Prober JM, et al. A system for rapid DNA sequencing with fluorescent chain-terminating dideoxynucleotides. Science. 1987;238:336. - PubMed
-
- NIH News Release, October 14, 2004, http://www.genome.gov/12513210
-
- Nyren P, Pettersson B, Uhlen M. Solid phase DNA minisequencing by an enzymatic luminometric inorganic pyrophosphate detection assay. Anal Biochem. 1993;208:171. - PubMed
-
- Ronaghi M, et al. Real-time DNA sequencing using detection of pyrophosphate release. Anal Biochem. 1996;242:84. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
