The p85 regulatory subunit of phosphoinositide 3-kinase down-regulates IRS-1 signaling via the formation of a sequestration complex
- PMID: 16043515
- PMCID: PMC2171479
- DOI: 10.1083/jcb.200503088
The p85 regulatory subunit of phosphoinositide 3-kinase down-regulates IRS-1 signaling via the formation of a sequestration complex
Abstract
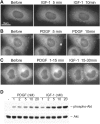
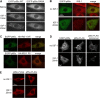
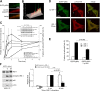
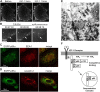
Phosphoinositide (PI) 3-kinase is required for most insulin and insulin-like growth factor (IGF) 1-dependent cellular responses. The p85 regulatory subunit of PI 3-kinase is required to mediate the insulin-dependent recruitment of PI 3-kinase to the plasma membrane, yet mice with reduced p85 expression have increased insulin sensitivity. To further understand the role of p85, we examined IGF-1-dependent translocation of p85alpha by using a green fluorescence protein (GFP)-tagged p85alpha (EGFP-p85alpha). In response to IGF-1, but not to PDGF signaling, EGFP-p85alpha translocates to discrete foci in the cell. These foci contain the insulin receptor substrate (IRS) 1 adaptor molecule, and their formation requires the binding of p85 to IRS-1. Surprisingly, monomeric p85 is preferentially localized to these foci compared with the p85-p110 dimer, and these foci are not sites of phosphatidylinositol-3,4,5-trisphosphate production. Ultrastructural analysis reveals that p85-IRS-1 foci are cytosolic protein complexes devoid of membrane. These results suggest a mechanism of signal down-regulation of IRS-1 that is mediated by monomeric p85 through the formation of a sequestration complex between p85 and IRS-1.
Figures

Similar articles
-
SH3 domain of the phosphatidylinositol 3-kinase regulatory subunit is responsible for the formation of a sequestration complex with insulin receptor substrate-1.Biochem Biophys Res Commun. 2008 Jan 18;365(3):433-8. doi: 10.1016/j.bbrc.2007.10.187. Epub 2007 Nov 6. Biochem Biophys Res Commun. 2008. PMID: 17991427
-
Interferon-dependent activation of the serine kinase PI 3'-kinase requires engagement of the IRS pathway but not the Stat pathway.Biochem Biophys Res Commun. 2000 Apr 2;270(1):158-62. doi: 10.1006/bbrc.2000.2402. Biochem Biophys Res Commun. 2000. PMID: 10733921
-
IRS-1 is a common element in insulin and insulin-like growth factor-I signaling to the phosphatidylinositol 3'-kinase.Endocrinology. 1993 Apr;132(4):1421-30. doi: 10.1210/endo.132.4.8384986. Endocrinology. 1993. PMID: 8384986
-
Class IA PI3K regulatory subunits: p110-independent roles and structures.Biochem Soc Trans. 2020 Aug 28;48(4):1397-1417. doi: 10.1042/BST20190845. Biochem Soc Trans. 2020. PMID: 32677674 Free PMC article. Review.
-
Structure and function of phosphatidylinositol-3,4 kinase.Cell Signal. 2000 Mar;12(3):135-42. doi: 10.1016/s0898-6568(99)00086-8. Cell Signal. 2000. PMID: 10704820 Review.
Cited by
-
mTOR pathway activation is a favorable prognostic factor in human prostate adenocarcinoma.Oncotarget. 2016 May 31;7(22):32916-24. doi: 10.18632/oncotarget.8767. Oncotarget. 2016. PMID: 27096957 Free PMC article.
-
Full-length adiponectin attenuates insulin signaling and inhibits insulin-stimulated amino Acid transport in human primary trophoblast cells.Diabetes. 2010 May;59(5):1161-70. doi: 10.2337/db09-0824. Epub 2010 Feb 11. Diabetes. 2010. PMID: 20150288 Free PMC article.
-
The regulatory subunits of PI3K, p85α and p85β, differentially affect BRD7-mediated regulation of insulin signaling.J Mol Cell Biol. 2022 Jan 29;13(12):889-901. doi: 10.1093/jmcb/mjab073. J Mol Cell Biol. 2022. PMID: 34751372 Free PMC article.
-
Demonstrated brain insulin resistance in Alzheimer's disease patients is associated with IGF-1 resistance, IRS-1 dysregulation, and cognitive decline.J Clin Invest. 2012 Apr;122(4):1316-38. doi: 10.1172/JCI59903. J Clin Invest. 2012. PMID: 22476197 Free PMC article.
-
Paradoxical dominant negative activity of an immunodeficiency-associated activating PIK3R1 variant.bioRxiv [Preprint]. 2024 Nov 5:2023.11.02.565250. doi: 10.1101/2023.11.02.565250. bioRxiv. 2024. Update in: Elife. 2025 Jan 21;13:RP94420. doi: 10.7554/eLife.94420. PMID: 38077044 Free PMC article. Updated. Preprint.
References
-
- Alessi, D.R., S.R. James, C.P. Downes, A.B. Holmes, P.R. Gaffney, C.B. Reese, and P. Cohen. 1997. Characterization of a 3-phosphoinositide-dependent protein kinase which phosphorylates and activates protein kinase Balpha. Curr. Biol. 7:261–269. - PubMed
-
- Barbour, L.A., J. Shao, L. Qiao, W. Leitner, M. Anderson, J.E. Friedman, and B. Draznin. 2004. Human placental growth hormone increases expression of the p85 regulatory unit of phosphatidylinositol 3-kinase and triggers severe insulin resistance in skeletal muscle. Endocrinology. 145:1144–1150. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

