Retrograde movement of tRNAs from the cytoplasm to the nucleus in Saccharomyces cerevisiae
- PMID: 16040803
- PMCID: PMC1183567
- DOI: 10.1073/pnas.0503836102
Retrograde movement of tRNAs from the cytoplasm to the nucleus in Saccharomyces cerevisiae
Abstract
In eukaryotes, tRNAs transcribed in the nucleus function in cytoplasmic protein synthesis. The Ran-GTP-binding exportin, Los1p/Xpo-t, and additional pathway(s) mediate tRNA transport to the cytoplasm. Although tRNA movement was thought to be unidirectional, recent reports that yeast precursor tRNA splicing occurs in the cytoplasm, whereas fully spliced tRNAs can reside in the nucleus, require that either the precursor tRNA splicing machinery or mature tRNAs move from the cytoplasm to the nucleus. Our data argue against the first possibility and strongly support the second. Combining heterokaryon analysis with fluorescence in situ hybridization, we show that a foreign tRNA encoded by one nucleus can move from the cytoplasm to a second nucleus that does not encode the tRNA. We also discovered nuclear accumulation of endogenous cytoplasmic tRNAs in haploid yeast cells in response to nutritional deprivation. Nuclear accumulation of cytoplasmic tRNA requires Ran and the Mtr10/Kap111 member of the importin-beta family. Retrograde tRNA nuclear import may provide a novel mechanism to regulate gene expression in eukaryotes.
Figures
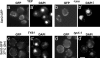
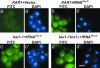
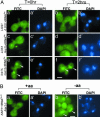
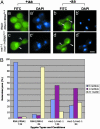
Comment in
-
Have tRNA, will travel.Proc Natl Acad Sci U S A. 2005 Aug 9;102(32):11127-8. doi: 10.1073/pnas.0504843102. Epub 2005 Aug 1. Proc Natl Acad Sci U S A. 2005. PMID: 16061803 Free PMC article. Review. No abstract available.
Similar articles
-
tRNA turnaround.Mol Cell. 2005 Aug 5;19(3):292-4. doi: 10.1016/j.molcel.2005.07.009. Mol Cell. 2005. PMID: 16061175
-
Retrograde transfer RNA nuclear import provides a new level of tRNA quality control in Saccharomyces cerevisiae.Proc Natl Acad Sci U S A. 2013 Dec 24;110(52):21042-7. doi: 10.1073/pnas.1316579110. Epub 2013 Dec 2. Proc Natl Acad Sci U S A. 2013. PMID: 24297920 Free PMC article.
-
Cex1p facilitates Rna1p-mediated dissociation of the Los1p-tRNA-Gsp1p-GTP export complex.Traffic. 2012 Feb;13(2):234-56. doi: 10.1111/j.1600-0854.2011.01304.x. Epub 2011 Nov 15. Traffic. 2012. PMID: 22008473
-
tRNA dynamics between the nucleus, cytoplasm and mitochondrial surface: Location, location, location.Biochim Biophys Acta Gene Regul Mech. 2018 Apr;1861(4):373-386. doi: 10.1016/j.bbagrm.2017.11.007. Epub 2017 Nov 28. Biochim Biophys Acta Gene Regul Mech. 2018. PMID: 29191733 Free PMC article. Review.
-
Review: transport of tRNA out of the nucleus-direct channeling to the ribosome?J Struct Biol. 2000 Apr;129(2-3):288-94. doi: 10.1006/jsbi.2000.4226. J Struct Biol. 2000. PMID: 10806079 Review.
Cited by
-
Transfer RNA post-transcriptional processing, turnover, and subcellular dynamics in the yeast Saccharomyces cerevisiae.Genetics. 2013 May;194(1):43-67. doi: 10.1534/genetics.112.147470. Genetics. 2013. PMID: 23633143 Free PMC article. Review.
-
Quality Control Pathways for Nucleus-Encoded Eukaryotic tRNA Biosynthesis and Subcellular Trafficking.Mol Cell Biol. 2015 Jun;35(12):2052-8. doi: 10.1128/MCB.00131-15. Epub 2015 Apr 6. Mol Cell Biol. 2015. PMID: 25848089 Free PMC article. Review.
-
Evolving specificity of tRNA 3-methyl-cytidine-32 (m3C32) modification: a subset of tRNAsSer requires N6-isopentenylation of A37.RNA. 2016 Sep;22(9):1400-10. doi: 10.1261/rna.056259.116. Epub 2016 Jun 27. RNA. 2016. PMID: 27354703 Free PMC article.
-
Export of Precursor tRNAIle from the Nucleus to the Cytoplasm in Human Cells.PLoS One. 2016 Apr 21;11(4):e0154044. doi: 10.1371/journal.pone.0154044. eCollection 2016. PLoS One. 2016. PMID: 27101286 Free PMC article.
-
Novel Links between TORC1 and Traditional Non-Coding RNA, tRNA.Genes (Basel). 2020 Aug 19;11(9):956. doi: 10.3390/genes11090956. Genes (Basel). 2020. PMID: 32825021 Free PMC article. Review.
References
-
- Hopper, A. K. & Phizicky, E. M. (2003) Genes Dev. 17, 162–180. - PubMed
-
- Lund, E. & Dahlberg, J. E. (1998) Science 282, 2082–2085. - PubMed
-
- Paushkin, S. V., Patel, M., Furia, B. S., Peltz, S. W. & Trotta, C. R. (2004) Cell 117, 311–321. - PubMed
-
- Huh, W. K., Falvo, J. V., Gerke, L. C., Carroll, A. S., Howson, R. W., Weissman, J. S. & O'Shea, E. K. (2003) Nature 425, 686–691. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

