The AML1-ETO fusion gene and the FLT3 length mutation collaborate in inducing acute leukemia in mice
- PMID: 16025155
- PMCID: PMC1174917
- DOI: 10.1172/JCI24225
The AML1-ETO fusion gene and the FLT3 length mutation collaborate in inducing acute leukemia in mice
Abstract
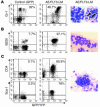
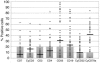
The molecular characterization of leukemia has demonstrated that genetic alterations in the leukemic clone frequently fall into 2 classes, those affecting transcription factors (e.g., AML1-ETO) and mutations affecting genes involved in signal transduction (e.g., activating mutations of FLT3 and KIT). This finding has favored a model of leukemogenesis in which the collaboration of these 2 classes of genetic alterations is necessary for the malignant transformation of hematopoietic progenitor cells. The model is supported by experimental data indicating that AML1-ETO and FLT3 length mutation (FLT3-LM), 2 of the most frequent genetic alterations in AML, are both insufficient on their own to cause leukemia in animal models. Here we report that AML1-ETO collaborates with FLT3-LM in inducing acute leukemia in a murine BM transplantation model. Moreover, in a series of 135 patients with AML1-ETO-positive AML, the most frequently identified class of additional mutations affected genes involved in signal transduction pathways including FLT3-LM or mutations of KIT and NRAS. These data support the concept of oncogenic cooperation between AML1-ETO and a class of activating mutations, recurrently found in patients with t(8;21), and provide a rationale for therapies targeting signal transduction pathways in AML1-ETO-positive leukemias.
Figures




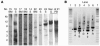
Similar articles
-
The ordered acquisition of Class II and Class I mutations directs formation of human t(8;21) acute myelogenous leukemia stem cell.Exp Hematol. 2014 Nov;42(11):955-65.e1-5. doi: 10.1016/j.exphem.2014.07.267. Epub 2014 Aug 4. Exp Hematol. 2014. PMID: 25101977
-
C-KIT mutation cooperates with full-length AML1-ETO to induce acute myeloid leukemia in mice.Proc Natl Acad Sci U S A. 2011 Feb 8;108(6):2450-5. doi: 10.1073/pnas.1019625108. Epub 2011 Jan 24. Proc Natl Acad Sci U S A. 2011. PMID: 21262832 Free PMC article.
-
AML1-ETO needs a partner: new insights into the pathogenesis of t(8;21) leukemia.Cell Cycle. 2005 Dec;4(12):1716-8. doi: 10.4161/cc.4.12.2256. Epub 2005 Dec 14. Cell Cycle. 2005. PMID: 16294039
-
Acute myeloid leukemia with the 8q22;21q22 translocation: secondary mutational events and alternative t(8;21) transcripts.Blood. 2007 Aug 1;110(3):799-805. doi: 10.1182/blood-2006-11-019265. Epub 2007 Apr 5. Blood. 2007. PMID: 17412887 Free PMC article. Review.
-
AML1-ETO driven acute leukemia: insights into pathogenesis and potential therapeutic approaches.Front Med. 2012 Sep;6(3):248-62. doi: 10.1007/s11684-012-0206-6. Epub 2012 Aug 9. Front Med. 2012. PMID: 22875638 Review.
Cited by
-
Profiling of somatic mutations and fusion genes in acute myeloid leukemia patients with FLT3-ITD or FLT3-TKD mutation at diagnosis reveals distinct evolutionary patterns.Exp Hematol Oncol. 2021 Apr 9;10(1):27. doi: 10.1186/s40164-021-00207-4. Exp Hematol Oncol. 2021. PMID: 33836835 Free PMC article.
-
Mouse models for core binding factor leukemia.Leukemia. 2015 Oct;29(10):1970-80. doi: 10.1038/leu.2015.181. Epub 2015 Jul 13. Leukemia. 2015. PMID: 26165235 Review.
-
FLT3-ITD cooperates with inv(16) to promote progression to acute myeloid leukemia.Blood. 2008 Feb 1;111(3):1567-74. doi: 10.1182/blood-2006-06-030312. Epub 2007 Oct 29. Blood. 2008. PMID: 17967943 Free PMC article.
-
The leukemogenicity of Hoxa9 depends on alternative splicing.Leukemia. 2014 Sep;28(9):1838-43. doi: 10.1038/leu.2014.74. Epub 2014 Feb 18. Leukemia. 2014. PMID: 24535405
-
CBFbeta is critical for AML1-ETO and TEL-AML1 activity.Blood. 2009 Mar 26;113(13):3070-9. doi: 10.1182/blood-2008-03-147207. Epub 2009 Jan 29. Blood. 2009. PMID: 19179469 Free PMC article.
References
-
- Look AT. Oncogenic transcription factors in the human acute leukemias. Science. 1997;278:1059–1064. - PubMed
-
- Rowley JD. The role of chromosome translocations in leukemogenesis. Semin. Hematol. 1999;36:59–72. - PubMed
-
- Gilliland DG, Tallman MS. Focus on acute leukemias. Cancer Cell. 2002;1:417–420. - PubMed
-
- Schnittger S, et al. Analysis of FLT3 length mutations in 1003 patients with acute myeloid leukemia: correlation to cytogenetics, FAB subtype, and prognosis in the AMLCG study and usefulness as a marker for the detection of minimal residual disease. Blood. 2002;100:59–66. - PubMed
-
- Gilliland DG. Molecular genetics of human leukemias: new insights into therapy [review] Semin. Hematol. 2002;39:6–11. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

