Amyloid ion channels: a common structural link for protein-misfolding disease
- PMID: 16020533
- PMCID: PMC1180768
- DOI: 10.1073/pnas.0502066102
Amyloid ion channels: a common structural link for protein-misfolding disease
Abstract
Protein conformational diseases, including Alzheimer's, Huntington's, and Parkinson's diseases, result from protein misfolding, giving a distinct fibrillar feature termed amyloid. Recent studies show that only the globular (not fibrillar) conformation of amyloid proteins is sufficient to induce cellular pathophysiology. However, the 3D structural conformations of these globular structures, a key missing link in designing effective prevention and treatment, remain undefined as of yet. By using atomic force microscopy, circular dichroism, gel electrophoresis, and electrophysiological recordings, we show here that an array of amyloid molecules, including amyloid-beta(1-40), alpha-synuclein, ABri, ADan, serum amyloid A, and amylin undergo supramolecular conformational change. In reconstituted membranes, they form morphologically compatible ion-channel-like structures and elicit single ion-channel currents. These ion channels would destabilize cellular ionic homeostasis and hence induce cell pathophysiology and degeneration in amyloid diseases.
Figures

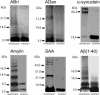
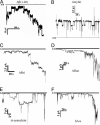
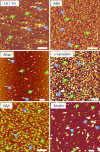


Similar articles
-
Amyloid beta protein forms ion channels: implications for Alzheimer's disease pathophysiology.FASEB J. 2001 Nov;15(13):2433-44. doi: 10.1096/fj.01-0377com. FASEB J. 2001. PMID: 11689468
-
Protein aggregation and deposition: implications for ion channel formation and membrane damage.Croat Med J. 2001 Aug;42(4):359-74. Croat Med J. 2001. PMID: 11471187 Review.
-
Amyloid beta ion channel: 3D structure and relevance to amyloid channel paradigm.Biochim Biophys Acta. 2007 Aug;1768(8):1966-75. doi: 10.1016/j.bbamem.2007.04.021. Epub 2007 May 3. Biochim Biophys Acta. 2007. PMID: 17553456 Free PMC article. Review.
-
Protein denaturation and aggregation: Cellular responses to denatured and aggregated proteins.Ann N Y Acad Sci. 2005 Dec;1066:181-221. doi: 10.1196/annals.1363.030. Ann N Y Acad Sci. 2005. PMID: 16533927 Review.
-
Conformational targeting of fibrillar polyglutamine proteins in live cells escalates aggregation and cytotoxicity.PLoS One. 2009 May 28;4(5):e5727. doi: 10.1371/journal.pone.0005727. PLoS One. 2009. PMID: 19492089 Free PMC article.
Cited by
-
Unveiling the Potential of Polyphenols as Anti-Amyloid Molecules in Alzheimer's Disease.Curr Neuropharmacol. 2023;21(4):787-807. doi: 10.2174/1570159X20666221010113812. Curr Neuropharmacol. 2023. PMID: 36221865 Free PMC article. Review.
-
Aggregation and fibril morphology of the Arctic mutation of Alzheimer's Aβ peptide by CD, TEM, STEM and in situ AFM.J Struct Biol. 2012 Oct;180(1):174-89. doi: 10.1016/j.jsb.2012.06.010. Epub 2012 Jun 28. J Struct Biol. 2012. PMID: 22750418 Free PMC article.
-
Versatile Structures of α-Synuclein.Front Mol Neurosci. 2016 Jun 20;9:48. doi: 10.3389/fnmol.2016.00048. eCollection 2016. Front Mol Neurosci. 2016. PMID: 27378848 Free PMC article. Review.
-
Conformational behavior and aggregation of ataxin-3 in SDS.PLoS One. 2013 Jul 22;8(7):e69416. doi: 10.1371/journal.pone.0069416. Print 2013. PLoS One. 2013. PMID: 23894474 Free PMC article.
-
Cytotoxic helix-rich oligomer formation by melittin and pancreatic polypeptide.PLoS One. 2015 Mar 24;10(3):e0120346. doi: 10.1371/journal.pone.0120346. eCollection 2015. PLoS One. 2015. PMID: 25803428 Free PMC article.
References
-
- Dobson, C. M. (2003) Nature 426, 884-890. - PubMed
-
- Selkoe, D. J. (2003) Nature 426, 900-904. - PubMed
-
- Revesz, T., Ghiso, J., Lashley, T., Plant, G., Rostagno, A., Frangione, B. & Holton, J. L. (2003) J. Neuropathol. Exp. Neurol. 62, 885-898. - PubMed
-
- Lin, H., Bhatia, R. & Lal, R. (2001) FASEB J. 15, 2433-2444. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

