Soluble mimetics of human immunodeficiency virus type 1 viral spikes produced by replacement of the native trimerization domain with a heterologous trimerization motif: characterization and ligand binding analysis
- PMID: 16014956
- PMCID: PMC1181572
- DOI: 10.1128/JVI.79.15.9954-9969.2005
Soluble mimetics of human immunodeficiency virus type 1 viral spikes produced by replacement of the native trimerization domain with a heterologous trimerization motif: characterization and ligand binding analysis
Abstract
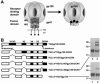
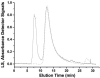
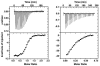
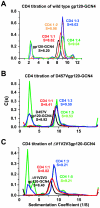
The human immunodeficiency virus type 1 (HIV-1) exterior envelope glycoprotein, gp120, mediates binding to the viral receptors and, along with the transmembrane glycoprotein gp41, is a major target for neutralizing antibodies. We asked whether replacing the gp41 fusion/trimerization domain with a stable trimerization motif might lead to a more stable gp120 trimer that would be amenable to structural and immunologic analysis. To obtain stable gp120 trimers, a heterologous trimerization motif, GCN4, was appended to the C terminus of YU2gp120. Biochemical analysis indicated that the gp120-GCN4 trimers were superior to gp140 molecules in their initial homogeneity, and trilobed structures were observable by electron microscopy. Biophysical analysis of gp120-GCN4 trimers by isothermal titration calorimetry (ITC) and ultracentrifugation analyses indicated that most likely two molecules of soluble CD4 could bind to one gp120-GCN4 trimer. To further examine restricted CD4 stoichiometric binding to the gp120-GCN4 trimers, we generated a low-affinity CD4 binding trimer by introducing a D457V change in the CD4 binding site of each gp120 monomeric subunit. The mutant trimers could definitively bind only one soluble CD4 molecule, as determined by ITC and sedimentation equilibrium centrifugation. These data indicate that there are weak interactions between the gp120 monomeric subunits of the GCN4-stabilized trimers that can be detected by low-affinity ligand sensing. By similar analysis, we also determined that removal of the variable loops V1, V2, and V3 in the context of the gp120-GCN4 proteins allowed the binding of three CD4 molecules per trimer. Interestingly, both the gp120-GCN4 variants displayed a restricted stoichiometry for the CD4-induced antibody 17b of one antibody molecule binding per trimer. This restriction was not evident upon removal of the variable loops V1 and V2 loops, consistent with conformational constraints in the wild-type gp120 trimers and similar to those inherent in the functional Env spike. Thus, the gp120-GCN4 trimers demonstrate several properties that are consistent with some of those anticipated for gp120 in the context of the viral spike.
Figures



Similar articles
-
Biochemically defined HIV-1 envelope glycoprotein variant immunogens display differential binding and neutralizing specificities to the CD4-binding site.J Biol Chem. 2012 Feb 17;287(8):5673-86. doi: 10.1074/jbc.M111.317776. Epub 2011 Dec 13. J Biol Chem. 2012. PMID: 22167180 Free PMC article.
-
Highly stable trimers formed by human immunodeficiency virus type 1 envelope glycoproteins fused with the trimeric motif of T4 bacteriophage fibritin.J Virol. 2002 May;76(9):4634-42. doi: 10.1128/jvi.76.9.4634-4642.2002. J Virol. 2002. PMID: 11932429 Free PMC article.
-
Effects of the I559P gp41 change on the conformation and function of the human immunodeficiency virus (HIV-1) membrane envelope glycoprotein trimer.PLoS One. 2015 Apr 7;10(4):e0122111. doi: 10.1371/journal.pone.0122111. eCollection 2015. PLoS One. 2015. PMID: 25849367 Free PMC article.
-
CD4 activation of HIV fusion.Int J Cell Cloning. 1992 Nov;10(6):323-32. doi: 10.1002/stem.5530100603. Int J Cell Cloning. 1992. PMID: 1281202 Review.
-
Structure-based design, synthesis and validation of CD4-mimetic small molecule inhibitors of HIV-1 entry: conversion of a viral entry agonist to an antagonist.Acc Chem Res. 2014 Apr 15;47(4):1228-37. doi: 10.1021/ar4002735. Epub 2014 Feb 6. Acc Chem Res. 2014. PMID: 24502450 Free PMC article. Review.
Cited by
-
Biochemically defined HIV-1 envelope glycoprotein variant immunogens display differential binding and neutralizing specificities to the CD4-binding site.J Biol Chem. 2012 Feb 17;287(8):5673-86. doi: 10.1074/jbc.M111.317776. Epub 2011 Dec 13. J Biol Chem. 2012. PMID: 22167180 Free PMC article.
-
Alterations in the immunogenic properties of soluble trimeric human immunodeficiency virus type 1 envelope proteins induced by deletion or heterologous substitutions of the V1 loop.J Virol. 2010 Oct;84(19):9932-46. doi: 10.1128/JVI.00868-10. Epub 2010 Jul 21. J Virol. 2010. PMID: 20660181 Free PMC article.
-
Stabilized HIV-1 envelope glycoprotein trimers lacking the V1V2 domain, obtained by virus evolution.J Biol Chem. 2010 Nov 19;285(47):36456-70. doi: 10.1074/jbc.M110.156588. Epub 2010 Sep 8. J Biol Chem. 2010. PMID: 20826824 Free PMC article.
-
Characterizing anti-HIV monoclonal antibodies and immune sera by defining the mechanism of neutralization.Hum Antibodies. 2005;14(3-4):101-13. Hum Antibodies. 2005. PMID: 16720980 Free PMC article.
-
Stoichiometric Analyses of Soluble CD4 to Native-like HIV-1 Envelope by Single-Molecule Fluorescence Spectroscopy.Cell Rep. 2019 Oct 1;29(1):176-186.e4. doi: 10.1016/j.celrep.2019.08.074. Cell Rep. 2019. PMID: 31577947 Free PMC article.
References
-
- Adis International Ltd. 2003. HIV gp120 vaccine - VaxGen: AIDSVAX, AIDSVAX B/B, AIDSVAX B/E, HIV gp120 vaccine - Genentech, HIV gp120 vaccine AIDSVAX - VaxGen, HIV vaccine AIDSVAX - VaxGen. Drugs R D 4:249-253. - PubMed
-
- Barnett, S. W., S. Rajasekar, H. Legg, B. Doe, D. H. Fuller, J. R. Haynes, C. M. Walker, and K. S. Steimer. 1997. Vaccination with HIV-1 gp120 DNA induces immune responses that are boosted by a recombinant gp120 protein subunit. Vaccine 15:869-873. - PubMed
-
- Belshe, R. B., G. J. Gorse, M. J. Mulligan, T. G. Evans, M. C. Keefer, J. L. Excler, A. M. Duliege, J. Tartaglia, W. I. Cox, J. McNamara, K. L. Hwang, A. Bradney, D. Montefiori, K. J. Weinhold, et al. 1998. Induction of immune responses to HIV-1 by canarypox virus (ALVAC) HIV-1 and gp120 SF-2 recombinant vaccines in uninfected volunteers. AIDS 12:2407-2415. - PubMed
-
- Berman, P. W., T. J. Gregory, L. Riddle, G. R. Nakamura, M. A. Champe, J. P. Porter, F. M. Wurm, R. D. Hershberg, E. K. Cobb, and J. W. Eichberg. 1990. Protection of chimpanzees from infection by HIV-1 after vaccination with recombinant glycoprotein gp120 but not gp160. Nature 345:622-625. - PubMed
-
- Binley, J. M., T. Wrin, B. Korber, M. B. Zwick, M. Wang, C. Chappey, G. Stiegler, R. Kunert, S. Zolla-Pazner, H. Katinger, C. J. Petropoulos, and D. R. Burton. 2004. Comprehensive cross-clade neutralization analysis of a panel of anti-human immunodeficiency virus type 1 monoclonal antibodies. J. Virol. 78:13232-13252. - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

