Kaposi's sarcoma-associated herpesvirus K-bZIP represses gene transcription via SUMO modification
- PMID: 16014952
- PMCID: PMC1181544
- DOI: 10.1128/JVI.79.15.9912-9925.2005
Kaposi's sarcoma-associated herpesvirus K-bZIP represses gene transcription via SUMO modification
Abstract
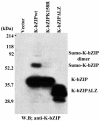
Kaposi's sarcoma-associated herpesvirus (KSHV) is a human gammaherpesvirus implicated in AIDS-related neoplasms. Previously, we demonstrated that the early lytic gene product K-bZIP is a transcriptional repressor that affects a subset of viral gene transcriptions mediated by the viral transactivator K-Rta (Y. Izumiya et al. J. Virol. 77:1441-1451, 2003). Sumoylation has emerged as an important posttranslational modification that affects the location and function of cellular and viral proteins and also plays a significant role in transcriptional repression along with Ubc9, the E2 SUMO conjugation enzyme. Here, we provide evidence that K-bZIP is sumoylated at the lysine 158 residue and associates with Ubc9 both in a cell-free system and in virus-infected BCBL-1 cells. Reporter assays showed that the expression of SUMO-specific protease 1 attenuated the transcriptional repression activity of K-bZIP. The expression of a K-bZIPK158R mutant, which was no longer sumoylated, exhibited the reduced transcriptional repression activity. This indicates that sumoylation plays an important part in the transcriptional repression activity of K-bZIP. Finally, chromatin immunoprecipitation experiments demonstrated that K-bZIP interacts with and recruits Ubc9 to specific KSHV promoters. Thus, our data indicate that K-bZIP is a SUMO adaptor, which recruits Ubc9 to specific viral target promoters, thereby exerting its transcriptional repression activity.
Figures



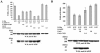



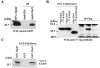

Similar articles
-
Modulation of global SUMOylation by Kaposi's sarcoma-associated herpesvirus and its effects on viral gene expression.J Med Virol. 2017 Nov;89(11):2011-2019. doi: 10.1002/jmv.24882. Epub 2017 Jul 21. J Med Virol. 2017. PMID: 28639696
-
Kaposi's sarcoma-associated herpesvirus K-bZIP is a coregulator of K-Rta: physical association and promoter-dependent transcriptional repression.J Virol. 2003 Jan;77(2):1441-51. doi: 10.1128/jvi.77.2.1441-1451.2003. J Virol. 2003. PMID: 12502859 Free PMC article.
-
K-bZIP of Kaposi's sarcoma-associated herpesvirus/human herpesvirus 8 (KSHV/HHV-8) binds KSHV/HHV-8 Rta and represses Rta-mediated transactivation.J Virol. 2003 Mar;77(6):3809-15. doi: 10.1128/jvi.77.6.3809-3815.2003. J Virol. 2003. PMID: 12610155 Free PMC article.
-
SUMO Ubc9 enzyme as a viral target.IUBMB Life. 2014 Jan;66(1):27-33. doi: 10.1002/iub.1240. Epub 2014 Jan 6. IUBMB Life. 2014. PMID: 24395713 Review.
-
SUMO and KSHV Replication.Cancers (Basel). 2014 Sep 29;6(4):1905-24. doi: 10.3390/cancers6041905. Cancers (Basel). 2014. PMID: 25268162 Free PMC article. Review.
Cited by
-
Sumoylation of the Epstein-Barr virus BZLF1 protein inhibits its transcriptional activity and is regulated by the virus-encoded protein kinase.J Virol. 2010 May;84(9):4383-94. doi: 10.1128/JVI.02369-09. Epub 2010 Feb 24. J Virol. 2010. PMID: 20181712 Free PMC article.
-
Mechanism of herpesvirus protein kinase UL13 in immune escape and viral replication.Front Immunol. 2022 Nov 30;13:1088690. doi: 10.3389/fimmu.2022.1088690. eCollection 2022. Front Immunol. 2022. PMID: 36531988 Free PMC article. Review.
-
Modification of papillomavirus E2 proteins by the small ubiquitin-like modifier family members (SUMOs).Virology. 2008 Sep 1;378(2):329-38. doi: 10.1016/j.virol.2008.06.008. Epub 2008 Jul 11. Virology. 2008. PMID: 18619639 Free PMC article.
-
Human pathogens and the host cell SUMOylation system.J Virol. 2012 Jan;86(2):642-54. doi: 10.1128/JVI.06227-11. Epub 2011 Nov 9. J Virol. 2012. PMID: 22072786 Free PMC article. Review.
-
Human Oncogenic Herpesvirus and Post-translational Modifications - Phosphorylation and SUMOylation.Front Microbiol. 2016 Jun 17;7:962. doi: 10.3389/fmicb.2016.00962. eCollection 2016. Front Microbiol. 2016. PMID: 27379086 Free PMC article. Review.
References
-
- Adamson, A. L., and S. C. Kenney. 1998. Rescue of the Epstein-Barr virus BZLF1 mutant, Z(S186A), early gene activation defect by the BRLF1 gene product. Virology 251:187-197. - PubMed
-
- AuCoin, D. P., K. S. Colletti, S. A. Cei, I. Papouskova, M. Tarrant, and G. S. Pari. 2003. Amplification of the Kaposi's sarcoma-associated herpesvirus/human herpesvirus 8 lytic origin of DNA replication is dependent upon a cis-acting AT-rich region and an ORF50 response element and the trans-acting factors ORF50 (K-Rta) and K8 (K-bZIP). Virology 318:542-555. - PubMed
-
- Chang, L. K., Y. H. Lee, T. S. Cheng, Y. R. Hong, P. J. Lu, J. J. Wang, W. H. Wang, C. W. Kuo, S. S. Li, and S. T. Liu. 2004. Post-translational modification of Rta of Epstein-Barr virus by SUMO-1. J. Biol. Chem. 279:38803-38812. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

