An atypical KdpD homologue from the cyanobacterium Anabaena sp. strain L-31: cloning, in vivo expression, and interaction with Escherichia coli KdpD-CTD
- PMID: 15995207
- PMCID: PMC1169523
- DOI: 10.1128/JB.187.14.4921-4927.2005
An atypical KdpD homologue from the cyanobacterium Anabaena sp. strain L-31: cloning, in vivo expression, and interaction with Escherichia coli KdpD-CTD
Abstract
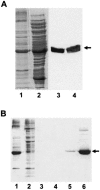
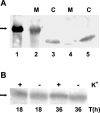
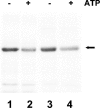
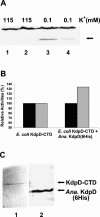
The kdpFABC operon of Escherichia coli, coding for the high-affinity K(+) transport system KdpFABC, is transcriptionally regulated by the products of the adjacently located kdpDE genes. The KdpD protein is a membrane-bound sensor kinase consisting of a large N-terminal domain and a C-terminal transmitter domain interconnected by four transmembrane segments (the transmembrane segments together with the C-terminal transmitter domain of KdpD are referred to as CTD), while KdpE is a cytosolic response regulator. We have cloned and sequenced the kdp operon from a nitrogen-fixing, filamentous cyanobacterium, Anabaena sp. strain L-31 (GenBank accession. number AF213466). The kdpABC genes are similar in size to those of E. coli, but the kdpD gene is short (coding only for 365 amino acids), showing homology only to the N-terminal domain of E. coli KdpD. A kdpE-like gene is absent in the vicinity of this operon. Anabaena KdpD with six C-terminal histidines was overproduced in E. coli and purified by Ni(2+)-nitrilotriacetic acid affinity chromatography. With antisera raised against the purified Anabaena KdpD, the protein was detected in Anabaena sp. strain L-31 membranes. The membrane-associated or soluble form of the Anabaena KdpD(6His) could be photoaffinity labeled with the ATP analog 8-azido-ATP, indicating the presence of an ATP binding site. The coproduction of Anabaena KdpD with E. coli KdpD-CTD decreased E. coli kdpFABC expression in response to K(+) limitation in vivo relative to the wild-type KdpD-CTD protein. In vitro experiments revealed that the kinase activity of the E. coli KdpD-CTD was unaffected, but its phosphatase activity increased in the presence of Anabaena KdpD(6His). To our knowledge this is the first report where a heterologous N-terminal domain (Anabaena KdpD) is shown to affect in trans KdpD-CTD (E. coli) activity, which is just opposite to that observed for the KdpD-N-terminal domain of E. coli.
Figures

Similar articles
-
Differential expression of the two kdp operons in the nitrogen-fixing cyanobacterium Anabaena sp. strain L-31.Appl Environ Microbiol. 2005 Sep;71(9):5297-303. doi: 10.1128/AEM.71.9.5297-5303.2005. Appl Environ Microbiol. 2005. PMID: 16151117 Free PMC article.
-
A chimeric Anabaena/ Escherichia coli KdpD protein (Anacoli KdpD) functionally interacts with E. coli KdpE and activates kdp expression in E. coli.Arch Microbiol. 2002 Aug;178(2):141-8. doi: 10.1007/s00203-002-0435-1. Epub 2002 May 29. Arch Microbiol. 2002. PMID: 12115059
-
The universal stress protein UspC scaffolds the KdpD/KdpE signaling cascade of Escherichia coli under salt stress.J Mol Biol. 2009 Feb 13;386(1):134-48. doi: 10.1016/j.jmb.2008.12.007. Epub 2008 Dec 11. J Mol Biol. 2009. PMID: 19101563
-
Towards an understanding of the molecular mechanisms of stimulus perception and signal transduction by the KdpD/KdpE system of Escherichia coli.J Mol Microbiol Biotechnol. 2002 May;4(3):223-8. J Mol Microbiol Biotechnol. 2002. PMID: 11931551 Review.
-
The complexity of the 'simple' two-component system KdpD/KdpE in Escherichia coli.FEMS Microbiol Lett. 2010 Mar;304(2):97-106. doi: 10.1111/j.1574-6968.2010.01906.x. Epub 2010 Jan 20. FEMS Microbiol Lett. 2010. PMID: 20146748 Review.
Cited by
-
The Kdp-ATPase system and its regulation.J Biosci. 2007 Apr;32(3):559-68. doi: 10.1007/s12038-007-0055-7. J Biosci. 2007. PMID: 17536175 Review.
-
Differential expression of the two kdp operons in the nitrogen-fixing cyanobacterium Anabaena sp. strain L-31.Appl Environ Microbiol. 2005 Sep;71(9):5297-303. doi: 10.1128/AEM.71.9.5297-5303.2005. Appl Environ Microbiol. 2005. PMID: 16151117 Free PMC article.
-
Cyanobacterial two-component proteins: structure, diversity, distribution, and evolution.Microbiol Mol Biol Rev. 2006 Jun;70(2):472-509. doi: 10.1128/MMBR.00046-05. Microbiol Mol Biol Rev. 2006. PMID: 16760311 Free PMC article. Review.
References
-
- Altendorf, K., and W. Epstein. 1996. The Kdp-ATPase of Escherichia coli, p. 403-420. In A. G. Lee (ed.), Biomembranes, vol. 5: ATPases. JAI Press, London, England.
-
- Apte, S. K., and A. Alahari. 1994. Role of alkali cations (K+ and Na+) in cyanobacterial nitrogen fixation and adaptation to salinity and osmotic stress. Indian J. Biochem. Biophys. 31:267-279. - PubMed
-
- Apte, S. K., and R. Haselkorn. 1990. Cloning of salinity stress-induced genes from salt tolerant nitrogen-fixing cyanobacteruim Anabaena torulosa. Plant Mol. Biol. 15:723-733. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources

