Heterologous late-domain sequences have various abilities to promote budding of human immunodeficiency virus type 1
- PMID: 15994797
- PMCID: PMC1168796
- DOI: 10.1128/jvi.79.14.9038-9045.2005
Heterologous late-domain sequences have various abilities to promote budding of human immunodeficiency virus type 1
Abstract
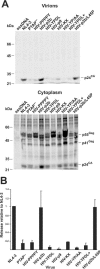
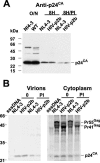
Retroviral late (L) domains present within Gag act in conjunction with cellular proteins to efficiently release virions from the surface of the cell. Three different critical core sequences have been identified as required elements for L-domain function: PPPY, PTAP (also PSAP), and YPDL, with different retroviruses utilizing one or two of these core sequences. The human immunodeficiency virus type 1 (HIV-1) L domain is centered around a PTAP sequence in the p6 region of Gag. To assess the ability of heterologous L-domain sequences to be functionally interchanged for those in full-length HIV-1, we produced a series of constructs that replaced PTAP-containing p6(Gag) sequences with those of PPPY- or YPDL-based L domains. While previous studies had found that L domains are interchangeable in other retroviruses, most of the sequences introduced into p6(Gag) failed to substitute for PTAP-mediated L-domain function. One exception was the 11-amino-acid p2b sequence of Rous sarcoma virus (RSV) Gag, which could fully restore HIV-1 budding, while a PPPPY sequence exchange alone did not. This suggests that the RSV L domain consists of more than simply its core L-domain sequence. The HIV-p2b chimera was as infectious as the wild type, produced normal virions, and was sensitive to proteasome inhibitors. These results show that L-domain sequences are not necessarily interchangeable. Thus, HIV-1 Gag might have a more stringent requirement for L-domain function than the other retroviruses previously studied.
Figures


Similar articles
-
Functional replacement and positional dependence of homologous and heterologous L domains in equine infectious anemia virus replication.J Virol. 2002 Feb;76(4):1569-77. doi: 10.1128/jvi.76.4.1569-1577.2002. J Virol. 2002. PMID: 11799151 Free PMC article.
-
Equine infectious anemia virus utilizes a YXXL motif within the late assembly domain of the Gag p9 protein.J Virol. 1997 Sep;71(9):6541-6. doi: 10.1128/JVI.71.9.6541-6546.1997. J Virol. 1997. PMID: 9261374 Free PMC article.
-
Both the PPPY and PTAP motifs are involved in human T-cell leukemia virus type 1 particle release.J Virol. 2004 Feb;78(3):1503-12. doi: 10.1128/jvi.78.3.1503-1512.2004. J Virol. 2004. PMID: 14722305 Free PMC article.
-
Late domain-dependent inhibition of equine infectious anemia virus budding.J Virol. 2004 Jan;78(2):724-32. doi: 10.1128/jvi.78.2.724-732.2004. J Virol. 2004. PMID: 14694104 Free PMC article.
-
Infectivity of Moloney murine leukemia virus defective in late assembly events is restored by late assembly domains of other retroviruses.J Virol. 2000 Aug;74(16):7250-60. doi: 10.1128/jvi.74.16.7250-7260.2000. J Virol. 2000. PMID: 10906179 Free PMC article.
Cited by
-
Proteomic and biochemical analysis of purified human immunodeficiency virus type 1 produced from infected monocyte-derived macrophages.J Virol. 2006 Sep;80(18):9039-52. doi: 10.1128/JVI.01013-06. J Virol. 2006. PMID: 16940516 Free PMC article.
-
Efficient production of HIV-1 virus-like particles from a mammalian expression vector requires the N-terminal capsid domain.PLoS One. 2011;6(11):e28314. doi: 10.1371/journal.pone.0028314. Epub 2011 Nov 30. PLoS One. 2011. PMID: 22140574 Free PMC article.
-
Mutations in human immunodeficiency virus type 1 nucleocapsid protein zinc fingers cause premature reverse transcription.J Virol. 2008 Oct;82(19):9318-28. doi: 10.1128/JVI.00583-08. Epub 2008 Jul 30. J Virol. 2008. PMID: 18667500 Free PMC article.
-
A PLPPV sequence in the p8 region of Gag provides late domain function for mouse mammary tumor virus.Virology. 2019 Sep;535:272-278. doi: 10.1016/j.virol.2019.07.015. Epub 2019 Jul 19. Virology. 2019. PMID: 31357166 Free PMC article.
-
The nucleocapsid region of human immunodeficiency virus type 1 Gag assists in the coordination of assembly and Gag processing: role for RNA-Gag binding in the early stages of assembly.J Virol. 2009 Aug;83(15):7718-27. doi: 10.1128/JVI.00099-09. Epub 2009 May 20. J Virol. 2009. PMID: 19457986 Free PMC article.
References
-
- Bleiber, G., S. Peters, R. Martinez, D. Cmarko, P. Meylan, and A. Telenti. 2004. The central region of human immunodeficiency virus type 1 p6 protein (Gag residues S14-I31) is dispensable for the virus in vitro. J. Gen. Virol. 85:921-927. - PubMed
-
- Blot, V., F. Perugi, B. Gay, M. C. Prevost, L. Briant, F. Tangy, H. Abriel, O. Staub, M. C. Dokhelar, and C. Pique. 2004. Nedd4.1-mediated ubiquitination and subsequent recruitment of Tsg101 ensure HTLV-1 Gag trafficking towards the multivesicular body pathway prior to virus budding. J. Cell Sci. 117:2357-2367. - PubMed
-
- Demirov, D. G., and E. O. Freed. 2004. Retrovirus budding. Virus Res. 106:87-102. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

