DNA damage induced hyperphosphorylation of replication protein A. 2. Characterization of DNA binding activity, protein interactions, and activity in DNA replication and repair
- PMID: 15938633
- PMCID: PMC4328999
- DOI: 10.1021/bi048057b
DNA damage induced hyperphosphorylation of replication protein A. 2. Characterization of DNA binding activity, protein interactions, and activity in DNA replication and repair
Abstract
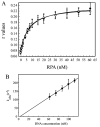
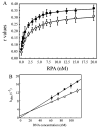
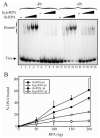
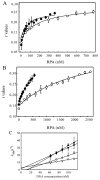
Replication protein A (RPA) is a heterotrimeric protein consisting of 70-, 34-, and 14- kDa subunits that is required for many DNA metabolic processes including DNA replication and DNA repair. Using a purified hyperphosphorylated form of RPA protein prepared in vitro, we have addressed the effects of hyperphosphorylation on steady-state and pre-steady-state DNA binding activity, the ability to support DNA repair and replication reactions, and the effect on the interaction with partner proteins. Equilibrium DNA binding activity measured by fluorescence polarization reveals no difference in ssDNA binding to pyrimidine-rich DNA sequences. However, RPA hyperphosphorylation results in a decreased affinity for purine-rich ssDNA and duplex DNA substrates. Pre-steady-state kinetic analysis is consistent with the equilibrium DNA binding and demonstrates a contribution from both the k(on) and k(off) to achieve these differences. The hyperphosphorylated form of RPA retains damage-specific DNA binding, and, importantly, the affinity of hyperphosphorylated RPA for damaged duplex DNA is 3-fold greater than the affinity of unmodified RPA for undamaged duplex DNA. The ability of hyperphosphorylated RPA to support DNA repair showed minor differences in the ability to support nucleotide excision repair (NER). Interestingly, under reaction conditions in which RPA is maintained in a hyperphosphorylated form, we also observed inhibition of in vitro DNA replication. Analyses of protein-protein interactions bear out the effects of hyperphosphorylated RPA on DNA metabolic pathways. Specifically, phosphorylation of RPA disrupts the interaction with DNA polymerase alpha but has no significant effect on the interaction with XPA. These results demonstrate that the effects of DNA damage induced hyperphosphorylation of RPA on DNA replication and DNA repair are mediated through alterations in DNA binding activity and protein-protein interactions.
Figures
Similar articles
-
Replication protein A interactions with DNA. III. Molecular basis of recognition of damaged DNA.Biochemistry. 2000 Feb 8;39(5):850-9. doi: 10.1021/bi991704s. Biochemistry. 2000. PMID: 10653628
-
Stopped-flow kinetic analysis of replication protein A-binding DNA: damage recognition and affinity for single-stranded DNA reveal differential contributions of k(on) and k(off) rate constants.J Biol Chem. 2001 Jun 22;276(25):22630-7. doi: 10.1074/jbc.M010314200. Epub 2001 Mar 6. J Biol Chem. 2001. PMID: 11278662
-
Strand-specific binding of RPA and XPA to damaged duplex DNA.Biochemistry. 2002 Feb 19;41(7):2402-8. doi: 10.1021/bi0112863. Biochemistry. 2002. PMID: 11841234
-
Replication protein A phosphorylation and the cellular response to DNA damage.DNA Repair (Amst). 2004 Aug-Sep;3(8-9):1015-24. doi: 10.1016/j.dnarep.2004.03.028. DNA Repair (Amst). 2004. PMID: 15279788 Review.
-
Functions of human replication protein A (RPA): from DNA replication to DNA damage and stress responses.J Cell Physiol. 2006 Aug;208(2):267-73. doi: 10.1002/jcp.20622. J Cell Physiol. 2006. PMID: 16523492 Free PMC article. Review.
Cited by
-
Phosphorylated RPA recruits PALB2 to stalled DNA replication forks to facilitate fork recovery.J Cell Biol. 2014 Aug 18;206(4):493-507. doi: 10.1083/jcb.201404111. Epub 2014 Aug 11. J Cell Biol. 2014. PMID: 25113031 Free PMC article.
-
Competition for DNA binding between the genome protector replication protein A and the genome modifying APOBEC3 single-stranded DNA deaminases.Nucleic Acids Res. 2022 Nov 28;50(21):12039-12057. doi: 10.1093/nar/gkac1121. Nucleic Acids Res. 2022. PMID: 36444883 Free PMC article.
-
Saccharomyces cerevisiae replication protein A binds to single-stranded DNA in multiple salt-dependent modes.Biochemistry. 2006 Oct 3;45(39):11958-73. doi: 10.1021/bi060994r. Biochemistry. 2006. PMID: 17002295 Free PMC article.
-
Protein phosphatase 2A-dependent dephosphorylation of replication protein A is required for the repair of DNA breaks induced by replication stress.Mol Cell Biol. 2009 Nov;29(21):5696-709. doi: 10.1128/MCB.00191-09. Epub 2009 Aug 24. Mol Cell Biol. 2009. PMID: 19704001 Free PMC article.
-
Ionizing radiation-dependent and independent phosphorylation of the 32-kDa subunit of replication protein A during mitosis.Nucleic Acids Res. 2009 Oct;37(18):6028-41. doi: 10.1093/nar/gkp605. Epub 2009 Aug 11. Nucleic Acids Res. 2009. PMID: 19671522 Free PMC article.
References
-
- Wold MS. Replication protein A: a heterotrimeric, single-stranded DNA-binding protein required for eukaryotic DNA metabolism. Annu. Rev. Biochem. 1997;66:61–92. - PubMed
-
- Iftode C, Daniely Y, Borowiec J. Replication protein A (RPA): the eukaryotic SSB, Crit. Rev. Biochem. Mol. Biol. 1999;34:141–180. - PubMed
-
- Brosh RM, Orren DK, Nehlin JO, Ravn PH, Kenny MK, Machwe A, Bohr VA. Functional and physical interaction between WRN helicase and human replication protein A. J. Biol. Chem. 1999;274:18341–18350. - PubMed
-
- He Z, Henricksen LA, Wold MS, Ingles CJ. RPA involvement in the damage-recognition and incision steps of nucleotide excision repair. Nature. 1995;374:566–569. - PubMed
-
- Bohr VA, Cooper M, Orren D, Machwe A, Piotrowski J, Sommers J, Karmakar P, Brosh R. Werner syndrome protein: biochemical properties and functional interactions. Exp. Gerontol. 2000;35:695–702. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

