The macro domain is an ADP-ribose binding module
- PMID: 15902274
- PMCID: PMC1142602
- DOI: 10.1038/sj.emboj.7600664
The macro domain is an ADP-ribose binding module
Abstract
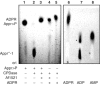
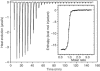
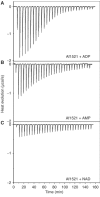
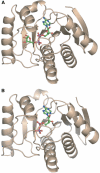
The ADP-ribosylation of proteins is an important post-translational modification that occurs in a variety of biological processes, including DNA repair, transcription, chromatin biology and long-term memory formation. Yet no protein modules are known that specifically recognize the ADP-ribose nucleotide. We provide biochemical and structural evidence that macro domains are high-affinity ADP-ribose binding modules. Our structural analysis reveals a conserved ligand binding pocket among the macro domain fold. Consistently, distinct human macro domains retain their ability to bind ADP-ribose. In addition, some macro domain proteins also recognize poly-ADP-ribose as a ligand. Our data suggest an important role for proteins containing macro domains in the biology of ADP-ribose.
Figures
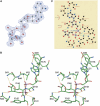

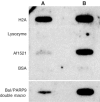
Similar articles
-
Differential activities of cellular and viral macro domain proteins in binding of ADP-ribose metabolites.J Mol Biol. 2009 Jan 9;385(1):212-25. doi: 10.1016/j.jmb.2008.10.045. Epub 2008 Nov 1. J Mol Biol. 2009. PMID: 18983849 Free PMC article.
-
Structural and functional basis for ADP-ribose and poly(ADP-ribose) binding by viral macro domains.J Virol. 2006 Sep;80(17):8493-502. doi: 10.1128/JVI.00713-06. J Virol. 2006. PMID: 16912299 Free PMC article.
-
The structure of ADP-ribose pyrophosphatase reveals the structural basis for the versatility of the Nudix family.Nat Struct Biol. 2001 May;8(5):467-72. doi: 10.1038/87647. Nat Struct Biol. 2001. PMID: 11323725
-
Players in ADP-ribosylation: Readers and Erasers.Curr Protein Pept Sci. 2016;17(7):654-667. doi: 10.2174/1389203717666160419144846. Curr Protein Pept Sci. 2016. PMID: 27090904 Review.
-
ADP-ribosylation of histones by ARTD1: an additional module of the histone code?FEBS Lett. 2011 Jun 6;585(11):1595-9. doi: 10.1016/j.febslet.2011.03.031. Epub 2011 Mar 21. FEBS Lett. 2011. PMID: 21420964 Review.
Cited by
-
A systematic analysis of the PARP protein family identifies new functions critical for cell physiology.Nat Commun. 2013;4:2240. doi: 10.1038/ncomms3240. Nat Commun. 2013. PMID: 23917125 Free PMC article.
-
ADP-ribose-1"-monophosphatase: a conserved coronavirus enzyme that is dispensable for viral replication in tissue culture.J Virol. 2005 Oct;79(20):12721-31. doi: 10.1128/JVI.79.20.12721-12731.2005. J Virol. 2005. PMID: 16188975 Free PMC article.
-
Dissection of the unusual structural and functional properties of the variant H2A.Bbd nucleosome.EMBO J. 2006 Sep 20;25(18):4234-44. doi: 10.1038/sj.emboj.7601310. Epub 2006 Sep 7. EMBO J. 2006. PMID: 16957777 Free PMC article.
-
Hepatitis E Virus Replication.Viruses. 2019 Aug 6;11(8):719. doi: 10.3390/v11080719. Viruses. 2019. PMID: 31390784 Free PMC article. Review.
-
PARP14 is a PARP with both ADP-ribosyl transferase and hydrolase activities.Sci Adv. 2023 Sep 15;9(37):eadi2687. doi: 10.1126/sciadv.adi2687. Epub 2023 Sep 13. Sci Adv. 2023. PMID: 37703374 Free PMC article.
References
-
- Abrahams JP, Leslie AGW, Lutter R, Walker JE (1994) Structure at 2.8 Angstrom resolution of the F1-ATPase from bovine heart mitochondria. Nature 370: 621–628 - PubMed
-
- Adamietz P, Rudolph A (1984) ADP-ribosylation of nuclear proteins in vivo. Identification of histone H2B as a major acceptor for mono- and poly(ADP-ribose) in dimethyl sulfate-treated hepatoma AH 7974 cells. J Biol Chem 259: 6841–6846 - PubMed
-
- Aguiar RC, Yakushijin Y, Kharbanda S, Salgia R, Fletcher JA, Shipp MA (2000) BAL is a novel risk-related gene in diffuse large B-cell lymphomas that enhances cellular migration. Blood 96: 4328–4334 - PubMed
-
- Allen MD, Buckle AM, Cordell SC, Lowe J, Bycroft M (2003) The crystal structure of AF1521 a protein from Archaeoglobus fulgidus with homology to the non-histone domain of macroH2A. J Mol Biol 330: 503–511 - PubMed
-
- Ame JC, Spenlehauer C, de Murcia G (2004) The PARP superfamily. Bioessays 26: 882–893 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

