Quantification of G protein Gaalphas subunit splice variants in different human tissues and cells using pyrosequencing
- PMID: 15892449
- PMCID: PMC6009110
- DOI: 10.3727/000000005783992124
Quantification of G protein Gaalphas subunit splice variants in different human tissues and cells using pyrosequencing
Abstract
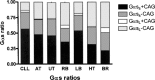
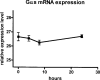
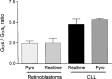
The G protein Galphas is derived from four alternatively spliced transcripts, two long variants (Galphas(L)+CAG and Galphas(L)-CAG), which include an extra 45-bp segment, and two short variants (Galphas(S)+CAG and Galphas(S)-CAG). The long and short forms differ in each case by splicing in or out of a serine residue encoded at the 3' end of the variable exon 3. The relative expression of all four variants in human tissues is poorly investigated due to experimental limitations. We therefore established a method for reliable relative mRNA quantification of these splice variants based on the Pyrosequencing technology, and determined Galphas transcript ratios in various human tissues and cells. Galphas(S)/Galphas ratio was highest in blood mononuclear cells (0.84 +/- 0.02, n = 16) and lowest in the brain (0.51 +/- 0.14, n = 3). The different ranges resulted from differences in Galphas(S)+CAG ratios, which ranged from a total Galphas ratio of 0.32 +/- 0.07 (n = 12) in heart tissue to 0.57 +/- 0.03 (n = 16) in blood mononuclear cells (p < 0.0001), whereas the Galphas(S)-CAG ratio was rather constant and ranged from 0.22 +/- 0.04 (n = 7) in retinoblastoma cells to 0.27 +/- 0.04 in lymphocytes (p = 0.19). The Galphas(L)+CAG ratio ranged from 0.02 +/- 0.02 in heart tissue to 0.05 +/- 0.01 in retinoblastoma cells, with a varying proportion of Galphas(L)-CAG, which ranged from 0.14 +/- 0.02 in blood mononuclear cells to 0.41 +/- 0.08 in heart tissue. Stimulation of immortalized B lymphoblasts with isoproterenol resulted in significant changes of splice variant ratios. Our data indicate that changes of long and short ratios of Galphas in different tissues affected Galphas(L)-CAG and Gas(S)+CAG rather than Galphas(L)+CAG and Galphas(S-)CAG. Furthermore, stimulation of cells seemed to affect splice variant ratios. These results are, therefore, suggestive of different biological functions of these variants.
Figures






Similar articles
-
Tissue specific expression of alternative splice forms of human cyclic nucleotide gated channel subunit CNGA3.Mol Vis. 2004 Oct 29;10:808-13. Mol Vis. 2004. PMID: 15534583
-
Agonist-induced release of splice variants of the alpha subunit of the stimulatory G-protein from rat cardiac membranes.Biochem Pharmacol. 1999 Mar 1;57(5):539-43. doi: 10.1016/s0006-2952(98)00335-9. Biochem Pharmacol. 1999. PMID: 9952317
-
Relative functions of Gαs and its extra-large variant XLαs in the endocrine system.Horm Metab Res. 2012 Sep;44(10):732-40. doi: 10.1055/s-0032-1316331. Epub 2012 Jun 22. Horm Metab Res. 2012. PMID: 22730013 Review.
-
Expression of alternatively spliced estrogen receptor alpha mRNAs is increased in breast cancer tissues.J Steroid Biochem Mol Biol. 2001 Nov;78(5):459-69. doi: 10.1016/s0960-0760(01)00118-2. J Steroid Biochem Mol Biol. 2001. PMID: 11738556
-
Mice maintain predominantly maternal Gαs expression throughout life in brown fat tissue (BAT), but not other tissues.Bone. 2017 Oct;103:177-187. doi: 10.1016/j.bone.2017.07.001. Epub 2017 Jul 8. Bone. 2017. PMID: 28694163 Free PMC article.
Cited by
-
Skeletal progenitors and the GNAS gene: fibrous dysplasia of bone read through stem cells.J Mol Endocrinol. 2010 Dec;45(6):355-64. doi: 10.1677/JME-10-0097. Epub 2010 Sep 14. J Mol Endocrinol. 2010. PMID: 20841428 Free PMC article. Review.
-
Analysis of splicing patterns by pyrosequencing.Nucleic Acids Res. 2009 Oct;37(19):e126. doi: 10.1093/nar/gkp626. Epub 2009 Aug 11. Nucleic Acids Res. 2009. PMID: 19671523 Free PMC article.
-
Violating the splicing rules: TG dinucleotides function as alternative 3' splice sites in U2-dependent introns.Genome Biol. 2007;8(8):R154. doi: 10.1186/gb-2007-8-8-r154. Genome Biol. 2007. PMID: 17672918 Free PMC article.
-
High resolution analysis of the human transcriptome: detection of extensive alternative splicing independent of transcriptional activity.BMC Genet. 2009 Oct 5;10:63. doi: 10.1186/1471-2156-10-63. BMC Genet. 2009. PMID: 19804644 Free PMC article.
-
Amelogenin sex determination by pyrosequencing of short PCR products.Int J Legal Med. 2008 Jul;122(4):333-5. doi: 10.1007/s00414-008-0228-4. Epub 2008 Mar 20. Int J Legal Med. 2008. PMID: 18351373
References
-
- Ahmadian A.; Gharizadeh B.; Gustafsson A. C.; Sterky F.; Nyren P.; Uhlen M.; Lundeberg J. Single-nucleotide polymorphism analysis by pyrosequencing. Anal. Biochem. 280(1):103–110; 2000. - PubMed
-
- Chaudhry A.; Granneman J. G. Developmental changes in adenylyl cyclase and GTP binding proteins in brown fat. Am. J. Physiol. 261(2 Pt. 2):R403–R411; 1991. - PubMed
-
- Cooper D. M.; Boyajian C. L.; Goldsmith P. K.; Unson C. G.; Spiegel A. Differential expression of low molecular weight form of Gs-alpha in neostriatum and cerebellum: Correlation with expression of calmodulin-independent adenylyl cyclase. Brain Res. 523(1):143–146; 1990. - PubMed
-
- Dobrev D.; Wettwer E.; Himmel H. M.; Kortner A.; Kuhlisch E.; Schuler S.; Siffert W.; Ravens U. G-Protein beta(3)-subunit 825T allele is associated with enhanced human atrial inward rectifier potassium currents. Circulation 102(6):692–697; 2000. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
