Elimination of ie1 significantly attenuates murine cytomegalovirus virulence but does not alter replicative capacity in cell culture
- PMID: 15890957
- PMCID: PMC1112098
- DOI: 10.1128/JVI.79.11.7182-7194.2005
Elimination of ie1 significantly attenuates murine cytomegalovirus virulence but does not alter replicative capacity in cell culture
Abstract
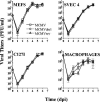
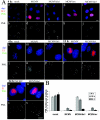
The major immediate-early (MIE) genes of cytomegaloviruses (CMV) are broadly thought to be decisive regulators of lytic replication and reactivation from latency. To directly assess the role of the MIE protein IE1 during the infection of murine CMV (MCMV), we constructed an MCMV with exon 4 of the ie1 gene deleted. We found that, independent of the multiplicity of infection, the resulting recombinant virus, MCMVdie1, which fails to express the IE1 protein, was fully competent for early gene expression and replicated in different cultured cell types with identical kinetics to those of parental or revertant virus. Immunofluorescence microscopy studies revealed that MCMVdie1 was greatly impaired in its capacity to disrupt promyelocytic leukemia bodies in NIH 3T3 cells early after infection, a process that has been proposed to increase viral transcription efficiency. We examined MCMVdie1 in the murine model using both immunocompetent BALB/c and severe combined immunodeficient (SCID) mice. When MCMVdie1 was inoculated into these two types of mice, significantly lower viral titers were detected in infected organs than in those of the wild-type virus-infected animals. Moreover, the ie1-deficient MCMV exhibited a markedly reduced virulence. While all animals infected with 5 x 10(4) PFU of parental virus died by 30 days postinfection, SCID mice infected with a similar dose of MCMVdie1 did not succumb before 60 days postinfection. The in vivo defective growth phenotype of MCMVdie1 was abrogated upon rescue of ie1. These results demonstrate the significance of the ie1 gene for promoting an acute MCMV infection and virulence yet indicate that MCMV is able to grow in vivo, although impaired, in the absence of the ie1 gene.
Figures




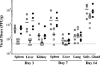

Similar articles
-
CD8 T cells control cytomegalovirus latency by epitope-specific sensing of transcriptional reactivation.J Virol. 2006 Nov;80(21):10436-56. doi: 10.1128/JVI.01248-06. Epub 2006 Aug 23. J Virol. 2006. PMID: 16928768 Free PMC article.
-
Murine cytomegalovirus with a deletion of genes spanning HindIII-J and -I displays altered cell and tissue tropism.J Virol. 1996 Mar;70(3):1365-74. doi: 10.1128/JVI.70.3.1365-1374.1996. J Virol. 1996. PMID: 8627652 Free PMC article.
-
Disruption of PML-associated nuclear bodies by IE1 correlates with efficient early stages of viral gene expression and DNA replication in human cytomegalovirus infection.Virology. 2000 Aug 15;274(1):39-55. doi: 10.1006/viro.2000.0448. Virology. 2000. PMID: 10936087
-
Genetic analyses of gene function and pathogenesis of murine cytomegalovirus by transposon-mediated mutagenesis.J Clin Virol. 2002 Aug;25 Suppl 2:S111-22. doi: 10.1016/s1386-6532(02)00096-3. J Clin Virol. 2002. PMID: 12361762 Review.
-
Phenotypes of major immediate-early gene mutants of mouse cytomegalovirus.Med Microbiol Immunol. 2008 Jun;197(2):233-40. doi: 10.1007/s00430-008-0076-3. Epub 2008 Feb 1. Med Microbiol Immunol. 2008. PMID: 18239940 Review.
Cited by
-
Viral latency drives 'memory inflation': a unifying hypothesis linking two hallmarks of cytomegalovirus infection.Med Microbiol Immunol. 2012 Nov;201(4):551-66. doi: 10.1007/s00430-012-0273-y. Epub 2012 Sep 19. Med Microbiol Immunol. 2012. PMID: 22991040 Review.
-
A short cis-acting motif in the M112-113 promoter region is essential for IE3 to activate M112-113 gene expression and is important for murine cytomegalovirus replication.J Virol. 2013 Mar;87(5):2639-47. doi: 10.1128/JVI.03171-12. Epub 2012 Dec 19. J Virol. 2013. PMID: 23255797 Free PMC article.
-
Ablation of the regulatory IE1 protein of murine cytomegalovirus alters in vivo pro-inflammatory TNF-alpha production during acute infection.PLoS Pathog. 2012;8(8):e1002901. doi: 10.1371/journal.ppat.1002901. Epub 2012 Aug 30. PLoS Pathog. 2012. PMID: 22952450 Free PMC article.
-
Murine cytomegalovirus perturbs endosomal trafficking of major histocompatibility complex class I molecules in the early phase of infection.J Virol. 2010 Nov;84(21):11101-12. doi: 10.1128/JVI.00988-10. Epub 2010 Aug 18. J Virol. 2010. PMID: 20719942 Free PMC article.
-
CD8 T cells control cytomegalovirus latency by epitope-specific sensing of transcriptional reactivation.J Virol. 2006 Nov;80(21):10436-56. doi: 10.1128/JVI.01248-06. Epub 2006 Aug 23. J Virol. 2006. PMID: 16928768 Free PMC article.
References
-
- Ahn, J. H., and G. S. Hayward. 2000. Disruption of PML-associated nuclear bodies by IE1 correlates with efficient early stages of viral gene expression and DNA replication in human cytomegalovirus infection. Virology 274:39-55. - PubMed
-
- Balthesen, M., L. Dreher, P. Lucin, and M. J. Reddehase. 1994. The establishment of cytomegalovirus latency in organs is not linked to local virus production during primary infection. J. Gen. Virol. 75:2329-2336. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

