Relationship of DFG16 to the Rim101p pH response pathway in Saccharomyces cerevisiae and Candida albicans
- PMID: 15879523
- PMCID: PMC1140096
- DOI: 10.1128/EC.4.5.890-899.2005
Relationship of DFG16 to the Rim101p pH response pathway in Saccharomyces cerevisiae and Candida albicans
Abstract
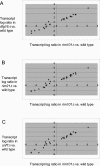
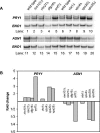
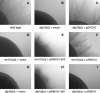
Many fungal pH responses depend upon conserved Rim101p/PacC transcription factors, which are activated by C-terminal proteolytic processing. The means by which environmental pH is sensed by this pathway are not known. Here, we report a screen of the Saccharomyces cerevisiae viable deletion mutant library that has yielded a new gene required for processed Rim101p accumulation, DFG16. An S. cerevisiae dfg16Delta mutant expresses Rim101p-repressed genes at elevated levels. In addition, Candida albicans dfg16Delta/dfg16Delta mutants are defective in alkaline pH-induced filamentation, and their defect is suppressed by expression of truncated Rim101-405p. Thus, Dfg16p is a functionally conserved Rim101p pathway member. Many proteins required for processed Rim101p accumulation are members of the ESCRT complex, which functions in the formation of multivesicular bodies (MVBs). Staining with the dye FM4-64 indicates that the S. cerevisiae dfg16Delta mutant does not have an MVB defect. We find that two transcripts, PRY1 and ASN1, respond to mutations that affect both the Rim101p and MVB pathways but not to mutations that affect only one pathway. The S. cerevisiae dfg16Delta mutation does not affect PRY1 and ASN1 expression, thus confirming that Dfg16p function is restricted to the Rim101p pathway. Dfg16p is homologous to Aspergillus nidulans PalH, a component of the well-characterized PacC processing pathway. We verify that the previously recognized PalH homolog, Rim21p, also functions in the S. cerevisiae Rim101p pathway. Dfg16p is predicted to have seven membrane-spanning segments and a long hydrophilic C-terminal region, as expected if Dfg16p were a G-protein-coupled receptor.
Figures



Similar articles
-
Multivesicular body-ESCRT components function in pH response regulation in Saccharomyces cerevisiae and Candida albicans.Mol Biol Cell. 2004 Dec;15(12):5528-37. doi: 10.1091/mbc.e04-08-0666. Epub 2004 Sep 15. Mol Biol Cell. 2004. PMID: 15371534 Free PMC article.
-
Candida albicans Rim13p, a protease required for Rim101p processing at acidic and alkaline pHs.Eukaryot Cell. 2004 Jun;3(3):741-51. doi: 10.1128/EC.3.3.741-751.2004. Eukaryot Cell. 2004. PMID: 15189995 Free PMC article.
-
The transcription factor Rim101p governs ion tolerance and cell differentiation by direct repression of the regulatory genes NRG1 and SMP1 in Saccharomyces cerevisiae.Mol Cell Biol. 2003 Jan;23(2):677-86. doi: 10.1128/MCB.23.2.677-686.2003. Mol Cell Biol. 2003. PMID: 12509465 Free PMC article.
-
Adaptation to environmental pH in Candida albicans and its relation to pathogenesis.Curr Genet. 2003 Oct;44(1):1-7. doi: 10.1007/s00294-003-0415-2. Epub 2003 Jun 18. Curr Genet. 2003. PMID: 12819929 Review.
-
Ambient pH gene regulation in fungi: making connections.Trends Microbiol. 2008 Jun;16(6):291-300. doi: 10.1016/j.tim.2008.03.006. Epub 2008 May 3. Trends Microbiol. 2008. PMID: 18457952 Review.
Cited by
-
Functional genomics analysis of the Saccharomyces cerevisiae iron responsive transcription factor Aft1 reveals iron-independent functions.Genetics. 2010 Jul;185(3):1111-28. doi: 10.1534/genetics.110.117531. Epub 2010 May 3. Genetics. 2010. PMID: 20439772 Free PMC article.
-
Effect of sequence-directed nucleosome disruption on cell-type-specific repression by alpha2/Mcm1 in the yeast genome.Eukaryot Cell. 2006 Nov;5(11):1925-33. doi: 10.1128/EC.00105-06. Epub 2006 Sep 15. Eukaryot Cell. 2006. PMID: 16980406 Free PMC article.
-
pH signaling in human fungal pathogens: a new target for antifungal strategies.Eukaryot Cell. 2014 Mar;13(3):342-52. doi: 10.1128/EC.00313-13. Epub 2014 Jan 17. Eukaryot Cell. 2014. PMID: 24442891 Free PMC article. Review.
-
Endosomal Na+ (K+)/H+ exchanger Nhx1/Vps44 functions independently and downstream of multivesicular body formation.J Biol Chem. 2011 Dec 23;286(51):44067-44077. doi: 10.1074/jbc.M111.282319. Epub 2011 Oct 13. J Biol Chem. 2011. PMID: 21998311 Free PMC article.
-
Cryptococcal titan cell formation is regulated by G-protein signaling in response to multiple stimuli.Eukaryot Cell. 2011 Oct;10(10):1306-16. doi: 10.1128/EC.05179-11. Epub 2011 Aug 5. Eukaryot Cell. 2011. PMID: 21821718 Free PMC article.
References
-
- Bensen, E. S., S. J. Martin, M. Li, J. Berman, and D. A. Davis. 2004. Transcriptional profiling in Candida albicans reveals new adaptive responses to extracellular pH and functions for Rim101p. Mol. Microbiol. 54:1335-1351. - PubMed
-
- Bowers, K., J. Lottridge, S. B. Helliwell, L. M. Goldthwaite, J. P. Luzio, and T. H. Stevens. 2004. Protein-protein interactions of ESCRT complexes in the yeast Saccharomyces cerevisiae. Traffic 5:194-210. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

