PU.1 regulates the commitment of adult hematopoietic progenitors and restricts granulopoiesis
- PMID: 15867096
- PMCID: PMC2213186
- DOI: 10.1084/jem.20050075
PU.1 regulates the commitment of adult hematopoietic progenitors and restricts granulopoiesis
Abstract
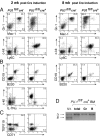
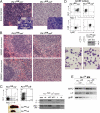
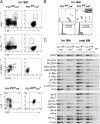
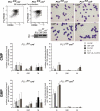
Although the transcription factor PU.1 is essential for fetal lymphomyelopoiesis, we unexpectedly found that elimination of the gene in adult mice allowed disturbed hematopoiesis, dominated by granulocyte production. Impaired production of lymphocytes was evident in PU.1-deficient bone marrow (BM), but myelocytes and clonogenic granulocytic progenitors that are responsive to granulocyte colony-stimulating factor or interleukin-3 increased dramatically. No identifiable common lymphoid or myeloid progenitor populations were discernable by flow cytometry; however, clonogenic assays suggested an overall increased frequency of blast colony-forming cells and BM chimeras revealed existence of long-term self-renewing PU.1-deficient cells that required PU.1 for lymphoid, but not granulocyte, generation. PU.1 deletion in granulocyte-macrophage progenitors, but not in common myeloid progenitors, resulted in excess granulocyte production; this suggested specific roles of PU.1 at different stages of myeloid development. These findings emphasize the distinct nature of adult hematopoiesis and reveal that PU.1 regulates the specification of the multipotent lymphoid and myeloid compartments and restrains, rather than promotes, granulopoiesis.
Figures


Similar articles
-
PU.1 regulates both cytokine-dependent proliferation and differentiation of granulocyte/macrophage progenitors.EMBO J. 1998 Aug 3;17(15):4456-68. doi: 10.1093/emboj/17.15.4456. EMBO J. 1998. PMID: 9687512 Free PMC article.
-
Transient expression of PU.1 commits multipotent progenitors to a myeloid fate whereas continued expression favors macrophage over granulocyte differentiation.Exp Hematol. 2003 Jan;31(1):39-47. doi: 10.1016/s0301-472x(02)01017-2. Exp Hematol. 2003. PMID: 12543105
-
Regulation of macrophage and neutrophil cell fates by the PU.1:C/EBPalpha ratio and granulocyte colony-stimulating factor.Nat Immunol. 2003 Oct;4(10):1029-36. doi: 10.1038/ni973. Epub 2003 Sep 7. Nat Immunol. 2003. PMID: 12958595
-
Stem cell fate specification: role of master regulatory switch transcription factor PU.1 in differential hematopoiesis.Stem Cells Dev. 2005 Apr;14(2):140-52. doi: 10.1089/scd.2005.14.140. Stem Cells Dev. 2005. PMID: 15910240 Review.
-
The importance of PU.1 concentration in hematopoietic lineage commitment and maturation.Blood Cells Mol Dis. 2003 Sep-Oct;31(2):229-33. doi: 10.1016/s1079-9796(03)00152-9. Blood Cells Mol Dis. 2003. PMID: 12972030 Review.
Cited by
-
Aging is associated with functional and molecular changes in distinct hematopoietic stem cell subsets.Nat Commun. 2024 Sep 11;15(1):7966. doi: 10.1038/s41467-024-52318-1. Nat Commun. 2024. PMID: 39261515 Free PMC article.
-
Hematopoiesis and T-cell specification as a model developmental system.Immunol Rev. 2016 May;271(1):72-97. doi: 10.1111/imr.12417. Immunol Rev. 2016. PMID: 27088908 Free PMC article. Review.
-
The role of transcription factors in the guidance of granulopoiesis.Am J Blood Res. 2012;2(1):57-65. Epub 2012 Jan 1. Am J Blood Res. 2012. PMID: 22432088 Free PMC article.
-
Runx1 deletion or dominant inhibition reduces Cebpa transcription via conserved promoter and distal enhancer sites to favor monopoiesis over granulopoiesis.Blood. 2012 May 10;119(19):4408-18. doi: 10.1182/blood-2011-12-397091. Epub 2012 Mar 26. Blood. 2012. PMID: 22451420 Free PMC article.
-
Launching the T-cell-lineage developmental programme.Nat Rev Immunol. 2008 Jan;8(1):9-21. doi: 10.1038/nri2232. Nat Rev Immunol. 2008. PMID: 18097446 Free PMC article. Review.
References
-
- Kondo, M., A.J. Wagers, M.G. Manz, S.S. Prohaska, D.C. Scherer, G.F. Beilhack, J.A. Shizuru, and I.L. Weissman. 2003. Biology of hematopoietic stem cells and progenitors: implications for clinical application. Annu. Rev. Immunol. 21:759–806. - PubMed
-
- Akashi, K., D. Traver, T. Miyamoto, and I.L. Weissman. 2000. A clonogenic common myeloid progenitor that gives rise to all myeloid lineages. Nature. 404:193–197. - PubMed
-
- Kondo, M., I.L. Weissman, and K. Akashi. 1997. Identification of clonogenic common lymphoid progenitors in mouse bone marrow. Cell. 91:661–672. - PubMed
-
- Traver, D., T. Miyamoto, J. Christensen, J. Iwasaki-Arai, K. Akashi, and I.L. Weissman. 2001. Fetal liver myelopoiesis occurs through distinct, prospectively isolatable progenitor subsets. Blood. 98:627–635. - PubMed
-
- Mebius, R.E., T. Miyamoto, J. Christensen, J. Domen, T. Cupedo, I.L. Weissman, and K. Akashi. 2001. The fetal liver counterpart of adult common lymphoid progenitors gives rise to all lymphoid lineages, CD45+CD4+CD3− cells, as well as macrophages. J. Immunol. 166:6593–6601. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials

