The locus encompassing the latency-associated transcript of herpes simplex virus type 1 interferes with and delays interferon expression in productively infected neuroblastoma cells and trigeminal Ganglia of acutely infected mice
- PMID: 15858001
- PMCID: PMC1091686
- DOI: 10.1128/JVI.79.10.6162-6171.2005
The locus encompassing the latency-associated transcript of herpes simplex virus type 1 interferes with and delays interferon expression in productively infected neuroblastoma cells and trigeminal Ganglia of acutely infected mice
Abstract
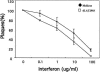
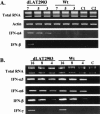
The herpes simplex virus type 1 (HSV-1) latency-associated transcript (LAT) is the only abundant viral transcript expressed in latently infected neurons. LAT inhibits apoptosis, suggesting that it regulates latency by promoting the survival of infected neurons. The LAT locus also contains a newly described gene (AL), which is antisense to LAT and partially overlaps LAT encoding sequences. When human (SK-N-SH) or mouse (neuro-2A) neuroblastoma cells were infected with a virus that does not express LAT or AL gene products (dLAT2903), beta interferon (IFN-beta) and IFN-alpha RNA expression was detected earlier relative to the same cells infected with HSV-1 strains that express LAT and AL. Infection of neuro-2A cells with dLAT2903 also led to higher levels of IFN-beta promoter activity than in cells infected with wild-type (wt) HSV-1. In contrast, IFN RNA expression was the same when human lung fibroblasts were infected with dLAT2903 or wt HSV-1. When BALB/c mice were infected with dLAT2903, IFN-alpha and IFN-beta RNA expression was readily detected in trigeminal ganglia (TG) 4 days after infection. These transcripts were not detected in TG of mice infected with wt HSV-1 or dLAT2903R (marker-rescued dLAT2903) until 6 days postinfection. When TG single-cell suspensions from infected BALB/c mice were prepared and incubated in vitro with wt HSV-1 as a source of antigen, TG cultures prepared from mice infected with dLAT2903 produced and secreted higher levels of IFN protein than wt HSV-1 or dLAT2903R. Collectively, these studies suggest that the LAT locus interferes with and delays IFN expression.
Figures





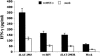
Similar articles
-
Herpes Simplex Virus Type 1 Preferentially Enhances Neuro-Inflammation and Senescence in Brainstem of Female Mice.J Virol. 2022 Sep 14;96(17):e0108122. doi: 10.1128/jvi.01081-22. Epub 2022 Aug 17. J Virol. 2022. PMID: 35975996 Free PMC article.
-
The persistent elevated cytokine mRNA levels in trigeminal ganglia of mice latently infected with HSV-1 are not due to the presence of latency associated transcript (LAT) RNAs.Virus Res. 1998 Mar;54(1):1-8. doi: 10.1016/s0168-1702(98)00007-0. Virus Res. 1998. PMID: 9660066
-
Infection of BALB/c mice with a herpes simplex virus type 1 recombinant virus expressing IFN-gamma driven by the LAT promoter.Virology. 2002 Oct 10;302(1):144-54. doi: 10.1006/viro.2002.1609. Virology. 2002. PMID: 12429523
-
Herpes simplex virus type 1 and bovine herpesvirus 1 latency.Clin Microbiol Rev. 2003 Jan;16(1):79-95. doi: 10.1128/CMR.16.1.79-95.2003. Clin Microbiol Rev. 2003. PMID: 12525426 Free PMC article. Review.
-
A Journey through the Minefield of the Discovery and Characterization of Latency-Related RNA/Latency-Associated Transcript.Viruses. 2024 Sep 30;16(10):1562. doi: 10.3390/v16101562. Viruses. 2024. PMID: 39459896 Free PMC article. Review.
Cited by
-
Identification of herpes simplex virus type 1 proteins encoded within the first 1.5 kb of the latency-associated transcript.J Neurovirol. 2009 Sep;15(5-6):439-48. doi: 10.3109/13550280903296353. J Neurovirol. 2009. PMID: 20175695
-
A protein encoded by the bovine herpesvirus 1 latency-related gene interacts with specific cellular regulatory proteins, including CCAAT enhancer binding protein alpha.J Virol. 2007 Jan;81(1):59-67. doi: 10.1128/JVI.01171-06. Epub 2006 Sep 20. J Virol. 2007. PMID: 16987965 Free PMC article.
-
The herpes simplex virus type 1 latency-associated transcript inhibits phenotypic and functional maturation of dendritic cells.Viral Immunol. 2012 Jun;25(3):204-15. doi: 10.1089/vim.2011.0091. Epub 2012 Apr 18. Viral Immunol. 2012. PMID: 22512280 Free PMC article.
-
Herpes Simplex Virus Type 1 Preferentially Enhances Neuro-Inflammation and Senescence in Brainstem of Female Mice.J Virol. 2022 Sep 14;96(17):e0108122. doi: 10.1128/jvi.01081-22. Epub 2022 Aug 17. J Virol. 2022. PMID: 35975996 Free PMC article.
-
Decreased reactivation of a herpes simplex virus type 1 (HSV-1) latency-associated transcript (LAT) mutant using the in vivo mouse UV-B model of induced reactivation.J Neurovirol. 2015 Oct;21(5):508-17. doi: 10.1007/s13365-015-0348-9. Epub 2015 May 22. J Neurovirol. 2015. PMID: 26002839 Free PMC article.
References
-
- Arthur, J. L., R. Everett, I. Brierley, and S. Efstathiou. 1998. Disruption of the 5′ and 3′ splice sites flanking the major latency-associated transcripts of herpes simplex virus type 1: evidence for alternate splicing in lytic and latent infections. J. Gen. Virol. 79:107-116. - PubMed
-
- Aubert, M., and J. A. Blaho. 2001. Modulation of apoptosis during herpes simplex virus infection in human cells. Microbes Infect. 3:859-866. - PubMed
-
- Casciano, I., A. de Ambrosis, M. Croce, G. Pagnan, A. Di Vinci, G. Allermanni, B. Banelli, M. Ponzoni, M. Romani, and S. Ferrini. 2004. Expression of the caspase-8 gene in neuroblastoma cells is regulated through an essential interferon-sensitive response element (ISRE). Cell Death Differ. 11:131-134. - PubMed
-
- Chawla-Sarkar, M., D. J. Lindner, Y.-F. Liu, B. R. Williams, G. C. Sen, R. H. Silverman, and E. C. Borden. 2003. Apoptosis and interferon: role of interferon-stimulated genes as mediators of apoptosis. Apoptosis 8:237-249. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

