Promoter features related to tissue specificity as measured by Shannon entropy
- PMID: 15833120
- PMCID: PMC1088961
- DOI: 10.1186/gb-2005-6-4-r33
Promoter features related to tissue specificity as measured by Shannon entropy
Abstract
Background: The regulatory mechanisms underlying tissue specificity are a crucial part of the development and maintenance of multicellular organisms. A genome-wide analysis of promoters in the context of gene-expression patterns in tissue surveys provides a means of identifying the general principles for these mechanisms.
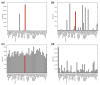
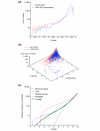
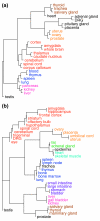
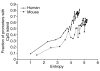
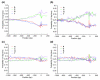
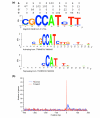
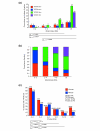
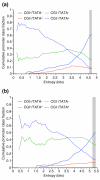
Results: We introduce a definition of tissue specificity based on Shannon entropy to rank human genes according to their overall tissue specificity and by their specificity to particular tissues. We apply our definition to microarray-based and expressed sequence tag (EST)-based expression data for human genes and use similar data for mouse genes to validate our results. We show that most genes show statistically significant tissue-dependent variations in expression level. We find that the most tissue-specific genes typically have a TATA box, no CpG island, and often code for extracellular proteins. As expected, CpG islands are found in most of the least tissue-specific genes, which often code for proteins located in the nucleus or mitochondrion. The class of genes with no CpG island or TATA box are the most common mid-specificity genes and commonly code for proteins located in a membrane. Sp1 was found to be a weak indicator of less-specific expression. YY1 binding sites, either as initiators or as downstream sites, were strongly associated with the least-specific genes.
Conclusions: We have begun to understand the components of promoters that distinguish tissue-specific from ubiquitous genes, to identify associations that can predict the broad class of gene expression from sequence data alone.
Figures
Similar articles
-
Genome-wide analysis of factors regulating gene expression in liver.Gene. 2007 Mar 15;389(2):114-21. doi: 10.1016/j.gene.2006.10.021. Epub 2006 Nov 7. Gene. 2007. PMID: 17174484
-
Clusters of regulatory signals for RNA polymerase II transcription associated with Alu family repeats and CpG islands in human promoters.Genomics. 2004 May;83(5):873-82. doi: 10.1016/j.ygeno.2003.11.001. Genomics. 2004. PMID: 15081116
-
Transcription from TATA-less promoters: dihydrofolate reductase as a model.Crit Rev Eukaryot Gene Expr. 1993;3(4):229-54. Crit Rev Eukaryot Gene Expr. 1993. PMID: 8286846 Review.
-
Dual promoter structure of ZFP106: regulation by myogenin and nuclear respiratory factor-1.Gene. 2005 Jan 3;344:143-59. doi: 10.1016/j.gene.2004.09.035. Epub 2004 Nov 19. Gene. 2005. PMID: 15656981
-
Promoter architecture and the evolvability of gene expression.J Biol. 2009;8(11):95. doi: 10.1186/jbiol204. Epub 2009 Dec 14. J Biol. 2009. PMID: 20017897 Free PMC article. Review.
Cited by
-
The unexpected traits associated with core promoter elements.Transcription. 2011 Sep-Oct;2(5):201-6. doi: 10.4161/trns.2.5.17271. Transcription. 2011. PMID: 22231114 Free PMC article. Review.
-
Shannon entropy as a metric for conditional gene expression in Neurospora crassa.G3 (Bethesda). 2021 Apr 15;11(4):jkab055. doi: 10.1093/g3journal/jkab055. G3 (Bethesda). 2021. PMID: 33751112 Free PMC article.
-
Engineering of a Histone-Recognition Domain in Dnmt3a Alters the Epigenetic Landscape and Phenotypic Features of Mouse ESCs.Mol Cell. 2015 Jul 2;59(1):89-103. doi: 10.1016/j.molcel.2015.05.017. Epub 2015 Jun 11. Mol Cell. 2015. PMID: 26073541 Free PMC article.
-
Testing the hypothesis of tissue selectivity: the intersection-union test and a Bayesian approach.Bioinformatics. 2009 Oct 1;25(19):2588-94. doi: 10.1093/bioinformatics/btp439. Epub 2009 Aug 11. Bioinformatics. 2009. PMID: 19671693 Free PMC article.
-
The cation efflux transporter ZnT8 (Slc30A8) is a major autoantigen in human type 1 diabetes.Proc Natl Acad Sci U S A. 2007 Oct 23;104(43):17040-5. doi: 10.1073/pnas.0705894104. Epub 2007 Oct 17. Proc Natl Acad Sci U S A. 2007. PMID: 17942684 Free PMC article.
References
-
- Bird AP. DNA methylation versus gene expression. J Embryol Exp Morphol. 1984;83(Suppl):31–40. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

