Use of adenoviral E1A protein to analyze K18 promoter deregulation in colon carcinoma cells discloses a role for CtBP1 and BRCA1
- PMID: 15831101
- PMCID: PMC1087485
- DOI: 10.1186/1471-2199-6-8
Use of adenoviral E1A protein to analyze K18 promoter deregulation in colon carcinoma cells discloses a role for CtBP1 and BRCA1
Abstract
Background: The promoter of the keratin 18 (K18) gene is 5- to 10-fold more active in tumorigenic (T-type) cell clones derived from the SW613-S human colon carcinoma cell line than in non-tumorigenic (NT-type) clones. We have reported previously that the mechanism responsible for this differential activity is acting on the minimal K18 promoter (TATA box and initiation site). This mechanism does not require the binding of a factor to a specific site on the DNA but involves the acetylation of a non-histone substrate. To get further insight into this mechanism, we investigated the effect of the adenovirus E1A protein on the activity of the K18 promoter, both in T and NT cells.
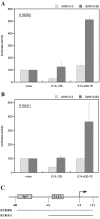
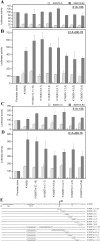
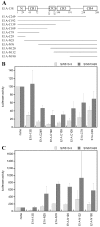
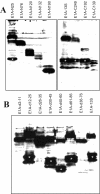
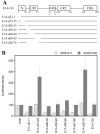
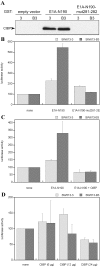
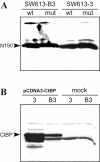
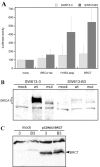
Results: Wild type adenovirus E1A protein and C-terminal deletion mutants inhibit the K18 promoter, specifically in T-type cells. The domain responsible for this inhibitory effect is located in the 12-25 region of the viral protein. E1A mutants that have lost this region but retain the PLDLS motif (the C-terminal binding site for CtBP1) stimulate the K18 promoter, specifically in NT cells. The inhibitory or stimulatory effects of the different E1A mutants are not dependent on a particular sequence of the promoter. An E1A N-terminal deletion mutant carrying point mutations in the PLDLS motif cannot stimulate the K18 promoter. CtBP1 interacts with CtIP, which is a known partner of BRCA1, itself a component of the RNA polymerase II holoenzyme. The stimulatory effect of two BRCA1 mutants, specifically in NT cells, implicates a tripartite BRCA1-CtIP-CtBP1 complex in the regulation of the K18 promoter.
Conclusion: Since we have shown previously that the K18 promoter is stimulated by deacetylase inhibitors, specifically in NT cells, we conclude that the activity of the promoter is repressed in NT cells by a mechanism involving the recruitment, by a BRCA1/CtIP complex, of CtBP1 and associated deacetylases to the preinitiation complex. We propose a model depicting the mechanism responsible for the differential activity of the K18 promoter between T and NT cells of the SW613-S cell line.
Figures
Similar articles
-
Transcriptional deregulation of the keratin 18 gene in human colon carcinoma cells results from an altered acetylation mechanism.Nucleic Acids Res. 2002 Aug 1;30(15):3312-22. doi: 10.1093/nar/gkf462. Nucleic Acids Res. 2002. PMID: 12140315 Free PMC article.
-
Transcriptional mechanisms responsible for the overexpression of the keratin 18 gene in cells of a human colon carcinoma cell line.Exp Cell Res. 1999 Apr 10;248(1):243-59. doi: 10.1006/excr.1999.4402. Exp Cell Res. 1999. PMID: 10094831
-
Impact of the interaction between adenovirus E1A and CtBP on host cell gene expression.Virus Res. 2005 Oct;113(1):51-63. doi: 10.1016/j.virusres.2005.04.015. Virus Res. 2005. PMID: 15899534
-
An Sp1 binding site and the minimal promoter contribute to overexpression of the cytokeratin 18 gene in tumorigenic clones relative to that in nontumorigenic clones of a human carcinoma cell line.Mol Cell Biol. 1995 May;15(5):2490-9. doi: 10.1128/MCB.15.5.2490. Mol Cell Biol. 1995. PMID: 7537848 Free PMC article.
-
Modulation of oncogenic transformation by the human adenovirus E1A C-terminal region.Curr Top Microbiol Immunol. 2004;273:139-61. doi: 10.1007/978-3-662-05599-1_5. Curr Top Microbiol Immunol. 2004. PMID: 14674601 Review.
Cited by
-
Oxaliplatin induces different cellular and molecular chemoresistance patterns in colorectal cancer cell lines of identical origins.BMC Genomics. 2013 Jul 16;14:480. doi: 10.1186/1471-2164-14-480. BMC Genomics. 2013. PMID: 23865481 Free PMC article.
-
The relationship between keratin 18 and epithelial-derived tumors: as a diagnostic marker, prognostic marker, and its role in tumorigenesis.Front Oncol. 2024 Oct 22;14:1445978. doi: 10.3389/fonc.2024.1445978. eCollection 2024. Front Oncol. 2024. PMID: 39502314 Free PMC article. Review.
References
-
- Ben Israel H, Kleinberger T. Adenovirus and cell cycle control. Front Biosci. 2002;7:d1369–d1395. - PubMed
-
- Schaeper U, Boyd JM, Verma S, Uhlmann E, Subramanian T, Chinnadurai G. Molecular cloning and characterization of a cellular phosphoprotein that interacts with a conserved C-terminal domain of adenovirus E1A involved in negative modulation of oncogenic transformation. Proc Natl Acad Sci U S A. 1995;92:10467–10471. - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

