Novel variants of human SCaMC-3, an isoform of the ATP-Mg/P(i) mitochondrial carrier, generated by alternative splicing from 3'-flanking transposable elements
- PMID: 15801905
- PMCID: PMC1180714
- DOI: 10.1042/BJ20050283
Novel variants of human SCaMC-3, an isoform of the ATP-Mg/P(i) mitochondrial carrier, generated by alternative splicing from 3'-flanking transposable elements
Abstract
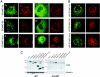
CaMCs (calcium-dependent mitochondrial carriers) represent a novel subfamily of metabolite carriers of mitochondria. The ATP-Mg/P(i) co-transporter, functionally characterized more than 20 years ago, has been identified to be a CaMC member. There are three isoforms of the ATP-Mg/P(i) carrier in mammals, SCaMC-1 (short CaMC-1), -2 and -3 (or APC-1, -3 and -2 respectively), corresponding to the genes SLC25A24, SLC25A25 and SLC25A23 respectively, as well as six N-terminal variants generated by alternative splicing for SCaMC-1 and -2 isoforms. In the present study, we describe four new variants of human SCaMC-3 generated by alternative splicing. The new mRNAs use the exon 9 3'-donor site and distinct 5'-acceptor sites from repetitive elements, in regions downstream of exon 10, the last exon in all SCaMCs. Transcripts lacking exon 10 (SCaMC-3b, -3b', -3c and -3d) code for shortened proteins lacking the last transmembrane domain of 422, 456 and 435 amino acids, and were found in human tissues and HEK-293T cells. Mitochondrial targeting of overexpressed SCaMC-3 variants is incomplete. Surprisingly, the import impairment is overcome by removing the N-terminal extension of these proteins, suggesting that the hydrophilic N-terminal domain also participates in the mitochondrial import process, as shown for the CaMC members aralar and citrin [Roesch, Hynds, Varga, Tranebjaerg and Koehler (2004) Hum. Mol. Genet. 13, 2101-2111].
Figures

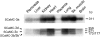

Similar articles
-
Identification of a novel human subfamily of mitochondrial carriers with calcium-binding domains.J Biol Chem. 2004 Jun 4;279(23):24701-13. doi: 10.1074/jbc.M401417200. Epub 2004 Mar 30. J Biol Chem. 2004. PMID: 15054102
-
Cellular expression and alternative splicing of SLC25A23, a member of the mitochondrial Ca2+-dependent solute carrier gene family.Gene. 2005 Jan 31;345(2):173-82. doi: 10.1016/j.gene.2004.11.028. Epub 2005 Jan 7. Gene. 2005. PMID: 15716113
-
Ca2+-regulated mitochondrial carriers of ATP-Mg2+/Pi: Evolutionary insights in protozoans.Biochim Biophys Acta Mol Cell Res. 2021 Jun;1868(7):119038. doi: 10.1016/j.bbamcr.2021.119038. Epub 2021 Apr 9. Biochim Biophys Acta Mol Cell Res. 2021. PMID: 33839167
-
Glutamate excitotoxicity and Ca2+-regulation of respiration: Role of the Ca2+ activated mitochondrial transporters (CaMCs).Biochim Biophys Acta. 2016 Aug;1857(8):1158-1166. doi: 10.1016/j.bbabio.2016.04.003. Epub 2016 Apr 7. Biochim Biophys Acta. 2016. PMID: 27060251 Review.
-
Mitochondrial transporters as novel targets for intracellular calcium signaling.Physiol Rev. 2007 Jan;87(1):29-67. doi: 10.1152/physrev.00005.2006. Physiol Rev. 2007. PMID: 17237342 Review.
Cited by
-
Inactivation of the mitochondrial carrier SLC25A25 (ATP-Mg2+/Pi transporter) reduces physical endurance and metabolic efficiency in mice.J Biol Chem. 2011 Apr 1;286(13):11659-71. doi: 10.1074/jbc.M110.203000. Epub 2011 Feb 4. J Biol Chem. 2011. PMID: 21296886 Free PMC article.
-
SCaMC-1Like a member of the mitochondrial carrier (MC) family preferentially expressed in testis and localized in mitochondria and chromatoid body.PLoS One. 2012;7(7):e40470. doi: 10.1371/journal.pone.0040470. Epub 2012 Jul 6. PLoS One. 2012. PMID: 22792342 Free PMC article.
-
Altered Slc25 family gene expression as markers of mitochondrial dysfunction in brain regions under experimental mixed anxiety/depression-like disorder.BMC Neurosci. 2018 Dec 11;19(1):79. doi: 10.1186/s12868-018-0480-6. BMC Neurosci. 2018. PMID: 30537945 Free PMC article.
-
The calcium-dependent ATP-Mg/Pi mitochondrial carrier is a target of glucose-induced calcium signalling in Saccharomyces cerevisiae.Biochem J. 2005 Dec 15;392(Pt 3):537-44. doi: 10.1042/BJ20050806. Biochem J. 2005. PMID: 16111475 Free PMC article.
-
Adenine nucleotide transporters in organelles: novel genes and functions.Cell Mol Life Sci. 2011 Apr;68(7):1183-206. doi: 10.1007/s00018-010-0612-3. Epub 2011 Jan 5. Cell Mol Life Sci. 2011. PMID: 21207102 Free PMC article. Review.
References
-
- Walker J. E., Runswick M. J. The mitochondrial transport protein superfamily. J. Bioenerg. Biomembr. 1993;4:35–46. - PubMed
-
- Kunji E. R. The role and structure of mitochondrial carriers. FEBS Lett. 2004;564:239–244. - PubMed
-
- Palmieri F. The mitochondrial transporter family (SLC25): physiological and pathological implications. Pflugers Arch. 2004;447:689–709. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

