Posttranscriptional downregulation of c-IAP2 by the ubiquitin protein ligase c-IAP1 in vivo
- PMID: 15798218
- PMCID: PMC1069614
- DOI: 10.1128/MCB.25.8.3348-3356.2005
Posttranscriptional downregulation of c-IAP2 by the ubiquitin protein ligase c-IAP1 in vivo
Abstract
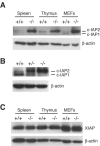
Inhibitor of apoptosis proteins (IAPs) c-IAP1 and c-IAP2 were identified as part of the tumor necrosis factor receptor 2 (TNFR2) signaling complex and have been implicated as intermediaries in tumor necrosis factor alpha signaling. Like all RING domain-containing IAPs, c-IAP1 and c-IAP2 have ubiquitin protein ligase (E3) activity. To explore the function of c-IAP1 in a physiologic setting, c-IAP1-deficient mice were generated by homologous gene recombination. These animals are viable and have no obvious sensitization to proapoptotic stimuli. Cells from c-IAP1(-/-) mice do, however, express markedly elevated levels of c-IAP2 protein in the absence of increased c-IAP2 mRNA. In contrast to reports implicating c-IAPs in the activation of NF-kappaB, resting and cytokine-induced NF-kappaB activation was not impaired in c-IAP1-deficient cells. Transient transfection studies with wild-type and E3-defective c-IAP1 revealed that c-IAP2 is a direct target for c-IAP1-mediated ubiquitination and subsequent degradation, which are potentiated by the adaptor function of TRAF2. Thus, the c-IAPs represent a pair of TNFR-associated ubiquitin protein ligases in which one regulates the expression of the other by a posttranscriptional and E3-dependent mechanism.
Figures
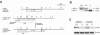

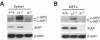



Similar articles
-
Ubiquitin binding modulates IAP antagonist-stimulated proteasomal degradation of c-IAP1 and c-IAP2(1).Biochem J. 2009 Jan 1;417(1):149-60. doi: 10.1042/BJ20081885. Biochem J. 2009. PMID: 18939944
-
c-IAP1 and c-IAP2 are critical mediators of tumor necrosis factor alpha (TNFalpha)-induced NF-kappaB activation.J Biol Chem. 2008 Sep 5;283(36):24295-9. doi: 10.1074/jbc.C800128200. Epub 2008 Jul 11. J Biol Chem. 2008. PMID: 18621737 Free PMC article.
-
TNF-RII and c-IAP1 mediate ubiquitination and degradation of TRAF2.Nature. 2002 Mar 21;416(6878):345-7. doi: 10.1038/416345a. Nature. 2002. PMID: 11907583
-
IAP family of cell death and signaling regulators.Methods Enzymol. 2014;545:35-65. doi: 10.1016/B978-0-12-801430-1.00002-0. Methods Enzymol. 2014. PMID: 25065885 Review.
-
(Un)expected roles of c-IAPs in apoptotic and NFkappaB signaling pathways.Cell Cycle. 2008 Jun 1;7(11):1511-21. doi: 10.4161/cc.7.11.5959. Epub 2008 Mar 16. Cell Cycle. 2008. PMID: 18469528 Review.
Cited by
-
Caspase 9 is constitutively activated in mouse oocytes and plays a key role in oocyte elimination during meiotic prophase progression.Dev Biol. 2013 May 1;377(1):213-23. doi: 10.1016/j.ydbio.2013.01.027. Epub 2013 Feb 4. Dev Biol. 2013. PMID: 23384561 Free PMC article.
-
cIAPs control RIPK1 kinase activity-dependent and -independent cell death and tissue inflammation.EMBO J. 2023 Nov 15;42(22):e113614. doi: 10.15252/embj.2023113614. Epub 2023 Oct 4. EMBO J. 2023. PMID: 37789765 Free PMC article.
-
RIP3: a molecular switch for necrosis and inflammation.Genes Dev. 2013 Aug 1;27(15):1640-9. doi: 10.1101/gad.223321.113. Genes Dev. 2013. PMID: 23913919 Free PMC article. Review.
-
Lycopene Inhibits Reactive Oxygen Species-Mediated NF-κB Signaling and Induces Apoptosis in Pancreatic Cancer Cells.Nutrients. 2019 Apr 1;11(4):762. doi: 10.3390/nu11040762. Nutrients. 2019. PMID: 30939781 Free PMC article.
-
Tumour-associated CD204+ microglia/macrophages accumulate in perivascular and perinecrotic niches and correlate with an interleukin-6-enriched inflammatory profile in glioblastoma.Neuropathol Appl Neurobiol. 2022 Feb;48(2):e12772. doi: 10.1111/nan.12772. Epub 2022 Jan 5. Neuropathol Appl Neurobiol. 2022. PMID: 34713474 Free PMC article.
References
-
- Abbondanzo, S. J., I. Gadi, and C. L. Stewart. 1993. Derivation of embryonic stem cell lines. Methods Enzymol. 225:803-823. - PubMed
-
- Akira, S., and T. Kishimoto. 1992. IL-6 and NF-IL6 in acute-phase response and viral infection. Immunol. Rev. 127:25-50. - PubMed
-
- Borden, K. L. 2000. RING domains: master builders of molecular scaffolds. J. Mol. Biol. 295:1103-1112. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous
