Role of the specific amino acid sequence of the membrane-spanning domain of human immunodeficiency virus type 1 in membrane fusion
- PMID: 15795258
- PMCID: PMC1069530
- DOI: 10.1128/JVI.79.8.4720-4729.2005
Role of the specific amino acid sequence of the membrane-spanning domain of human immunodeficiency virus type 1 in membrane fusion
Abstract
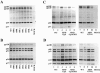
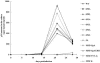
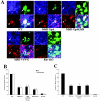
Fusion between cell and virus membranes mediated by gp41 initiates the life cycle of human immunodeficiency virus type 1. In contrast to the many studies that have elucidated the structure-function relationship of the ectodomain, the study of the membrane-spanning domain (MSD) has been rather limited. In particular, the role that the MSD's specific amino acid sequences may have in membrane fusion as well as other gp41 functions is not well understood. The MSD of gp41 contains well-conserved glycine residues that form the GXXXG motif (G, glycine; X, other amino acid residues), a motif often found at the helix-helix interface of membrane spanning alpha-helices. Here we examined the role that the specific amino acid sequence of the gp41 MSD has in gp41 function, particularly in membrane fusion, by making two types of MSD mutants: (i) glycine substitution mutants in which glycine residues of the MSD were mutated to alanine or leucine residues, and (ii) replacement mutants in which the entire MSD was replaced with one derived from glycophorin A or from vesicular stomatitis virus G. The substitution of glycines did not affect gp41 function. MSD-replacement mutants, however, showed severely impaired fusion activity. The assay using the Env expression vector revealed defects in membrane fusion after CD4 binding steps in the MSD-replacement mutants. In addition, the change in Env processing was noted for MSD-replacement mutants. These results suggest that the MSD of gp41 has a relatively wide but not unlimited tolerance for mutations and plays a critical role in membrane fusion as well as in other steps of Env biogenesis.
Figures

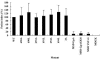

Similar articles
-
Mutations of conserved glycine residues within the membrane-spanning domain of human immunodeficiency virus type 1 gp41 can inhibit membrane fusion and incorporation of Env onto virions.Jpn J Infect Dis. 2006 Apr;59(2):77-84. Jpn J Infect Dis. 2006. PMID: 16632906
-
Role of the fusion peptide and membrane-proximal domain in HIV-1 envelope glycoprotein-mediated membrane fusion.Biochemistry. 2003 Dec 9;42(48):14150-8. doi: 10.1021/bi035154g. Biochemistry. 2003. PMID: 14640682
-
Role of envelope processing and gp41 membrane spanning domain in the formation of human immunodeficiency virus type 1 (HIV-1) fusion-competent envelope glycoprotein complex.Virus Res. 2007 Mar;124(1-2):103-12. doi: 10.1016/j.virusres.2006.10.009. Epub 2006 Nov 28. Virus Res. 2007. PMID: 17129629
-
The structural biology of type I viral membrane fusion.Nat Rev Mol Cell Biol. 2003 Apr;4(4):309-19. doi: 10.1038/nrm1076. Nat Rev Mol Cell Biol. 2003. PMID: 12671653 Review.
-
Evolutionary conservation of the membrane fusion machine.IUBMB Life. 1999 Aug;48(2):151-6. doi: 10.1080/713803503. IUBMB Life. 1999. PMID: 10794590 Review.
Cited by
-
Conformational changes of the HIV-1 envelope protein during membrane fusion are inhibited by the replacement of its membrane-spanning domain.J Biol Chem. 2010 May 7;285(19):14681-8. doi: 10.1074/jbc.M109.067090. Epub 2010 Mar 2. J Biol Chem. 2010. PMID: 20197275 Free PMC article.
-
Membrane interaction and structure of the transmembrane domain of influenza hemagglutinin and its fusion peptide complex.BMC Biol. 2008 Jan 15;6:2. doi: 10.1186/1741-7007-6-2. BMC Biol. 2008. PMID: 18197965 Free PMC article.
-
Role of the membrane-spanning domain of human immunodeficiency virus type 1 envelope glycoprotein in cell-cell fusion and virus infection.J Virol. 2008 Jun;82(11):5417-28. doi: 10.1128/JVI.02666-07. Epub 2008 Mar 19. J Virol. 2008. PMID: 18353944 Free PMC article.
-
The paramyxovirus fusion protein C-terminal region: mutagenesis indicates an indivisible protein unit.J Virol. 2012 Mar;86(5):2600-9. doi: 10.1128/JVI.06546-11. Epub 2011 Dec 14. J Virol. 2012. PMID: 22171273 Free PMC article.
-
Lipid interaction of the C terminus and association of the transmembrane segments facilitate atlastin-mediated homotypic endoplasmic reticulum fusion.Proc Natl Acad Sci U S A. 2012 Aug 7;109(32):E2146-54. doi: 10.1073/pnas.1208385109. Epub 2012 Jul 16. Proc Natl Acad Sci U S A. 2012. PMID: 22802620 Free PMC article.
References
-
- Aoki, Y., H. Aizaki, T. Shimoike, H. Tani, K. Ishii, I. Saito, Y. Matsuura, and T. Miyamura. 1998. A human liver cell line exhibits efficient translation of HCV RNAs produced by a recombinant adenovirus expressing T7 RNA polymerase. Virology 250:140-150. - PubMed
-
- Bullough, P. A., F. M. Hughson, J. J. Skehel, and D. C. Wiley. 1994. Structure of influenza haemagglutinin at the pH of membrane fusion. Nature 371:37-43. - PubMed
-
- Chan, D. C., D. Fass, J. M. Berger, and P. S. Kim. 1997. Core structure of gp41 from the HIV envelope glycoprotein. Cell 89:263-273. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

