A comprehensive update of the sequence and structure classification of kinases
- PMID: 15771780
- PMCID: PMC1079889
- DOI: 10.1186/1472-6807-5-6
A comprehensive update of the sequence and structure classification of kinases
Abstract
Background: A comprehensive update of the classification of all available kinases was carried out. This survey presents a complete global picture of this large functional class of proteins and confirms the soundness of our initial kinase classification scheme.
Results: The new survey found the total number of kinase sequences in the protein database has increased more than three-fold (from 17,310 to 59,402), and the number of determined kinase structures increased two-fold (from 359 to 702) in the past three years. However, the framework of the original two-tier classification scheme (in families and fold groups) remains sufficient to describe all available kinases. Overall, the kinase sequences were classified into 25 families of homologous proteins, wherein 22 families (approximately 98.8% of all sequences) for which three-dimensional structures are known fall into 10 fold groups. These fold groups not only include some of the most widely spread proteins folds, such as the Rossmann-like fold, ferredoxin-like fold, TIM-barrel fold, and antiparallel beta-barrel fold, but also all major classes (all alpha, all beta, alpha+beta, alpha/beta) of protein structures. Fold predictions are made for remaining kinase families without a close homolog with solved structure. We also highlight two novel kinase structural folds, riboflavin kinase and dihydroxyacetone kinase, which have recently been characterized. Two protein families previously annotated as kinases are removed from the classification based on new experimental data.
Conclusion: Structural annotations of all kinase families are now revealed, including fold descriptions for all globular kinases, making this the first large functional class of proteins with a comprehensive structural annotation. Potential uses for this classification include deduction of protein function, structural fold, or enzymatic mechanism of poorly studied or newly discovered kinases based on proteins in the same family.
Figures

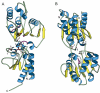

Similar articles
-
Sequence and structure classification of kinases.J Mol Biol. 2002 Jul 19;320(4):855-81. doi: 10.1016/s0022-2836(02)00538-7. J Mol Biol. 2002. PMID: 12095261
-
Classification of common functional loops of kinase super-families.Proteins. 2004 Aug 15;56(3):539-55. doi: 10.1002/prot.20136. Proteins. 2004. PMID: 15229886
-
Convergent evolution of similar enzymatic function on different protein folds: the hexokinase, ribokinase, and galactokinase families of sugar kinases.Protein Sci. 1993 Jan;2(1):31-40. doi: 10.1002/pro.5560020104. Protein Sci. 1993. PMID: 8382990 Free PMC article.
-
Phosphatidylinositol phosphate kinase: a link between protein kinase and glutathione synthase folds.J Mol Biol. 1999 Aug 13;291(2):239-47. doi: 10.1006/jmbi.1999.2973. J Mol Biol. 1999. PMID: 10438618 Review.
-
Protein kinases 6. The eukaryotic protein kinase superfamily: kinase (catalytic) domain structure and classification.FASEB J. 1995 May;9(8):576-96. FASEB J. 1995. PMID: 7768349 Review.
Cited by
-
Structure of the type III pantothenate kinase from Bacillus anthracis at 2.0 A resolution: implications for coenzyme A-dependent redox biology.Biochemistry. 2007 Mar 20;46(11):3234-45. doi: 10.1021/bi062299p. Epub 2007 Feb 27. Biochemistry. 2007. PMID: 17323930 Free PMC article.
-
ProCKSI: a decision support system for Protein (structure) Comparison, Knowledge, Similarity and Information.BMC Bioinformatics. 2007 Oct 26;8:416. doi: 10.1186/1471-2105-8-416. BMC Bioinformatics. 2007. PMID: 17963510 Free PMC article.
-
Molecular cloning and heterologous expression of the dehydrophos biosynthetic gene cluster.Chem Biol. 2010 Apr 23;17(4):402-11. doi: 10.1016/j.chembiol.2010.03.007. Chem Biol. 2010. PMID: 20416511 Free PMC article.
-
Diversity and versatility of the Thermotoga maritima sugar kinome.J Bacteriol. 2012 Oct;194(20):5552-63. doi: 10.1128/JB.01136-12. Epub 2012 Aug 10. J Bacteriol. 2012. PMID: 22885293 Free PMC article.
-
Fast and automated functional classification with MED-SuMo: an application on purine-binding proteins.Protein Sci. 2010 Apr;19(4):847-67. doi: 10.1002/pro.364. Protein Sci. 2010. PMID: 20162627 Free PMC article.
References
-
- Gerdes SY, Scholle MD, D'Souza M, Bernal A, Baev MV, Farrell M, Kurnasov OV, Daugherty MD, Mseeh F, Polanuyer BM, Campbell JW, Anantha S, Shatalin KY, Chowdhury SA, Fonstein MY, Osterman AL. From genetic footprinting to antimicrobial drug targets: examples in cofactor biosynthetic pathways. J Bacteriol. 2002;184:4555–4572. doi: 10.1128/JB.184.16.4555-4572.2002. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

