Replication-independent core histone dynamics at transcriptionally active loci in vivo
- PMID: 15769942
- PMCID: PMC1065721
- DOI: 10.1101/gad.1265205
Replication-independent core histone dynamics at transcriptionally active loci in vivo
Abstract
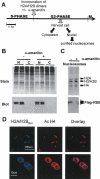
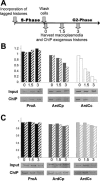
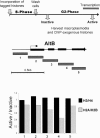
We used a novel labeling technique in the naturally synchronous organism Physarum polycephalum to examine the fate of core histones in G2 phase. We find rapid exchange of H2A/H2B dimers with free pools that is greatly diminished by treatment of the cells with alpha-amanitin. This exchange is enhanced in pol II-coding sequences compared with extragenic regions or inactive loci. In contrast, H3/H4 tetramers exhibit far lower levels of exchange in the pol II-transcribed genes tested, suggesting that tetramer exchange occurs via a distinct mechanism. However, we find that transcribed regions of the ribosomal RNA gene loci exhibit rapid exchange of H3/H4 tetramers. Thus, our data show that the majority of the pol II transcription-dependent histone exchange is due to elongation in vivo rather than promoter remodeling or other pol II-dependent alterations in promoter structure and, in contrast to pol I, pol II transcription through nucleosomes in vivo causes facile exchange of both H2A/H2B dimers while allowing conservation of epigenetic "marks" and other post-translational modifications on H3 and H4.
Figures
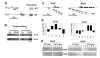
Similar articles
-
Histone dynamics during transcription: exchange of H2A/H2B dimers and H3/H4 tetramers during pol II elongation.Results Probl Cell Differ. 2006;41:77-90. doi: 10.1007/400_009. Results Probl Cell Differ. 2006. PMID: 16909891 Review.
-
Mechanism of histone survival during transcription by RNA polymerase II.Transcription. 2010 Sep-Oct;1(2):85-8. doi: 10.4161/trns.1.2.12519. Transcription. 2010. PMID: 21326897 Free PMC article.
-
Transcription-induced chromatin remodeling at the c-myc gene involves the local exchange of histone H2A.Z.J Biol Chem. 2005 Jul 1;280(26):25298-303. doi: 10.1074/jbc.M501784200. Epub 2005 May 6. J Biol Chem. 2005. PMID: 15878876
-
In vivo studies on the dynamics of histone-DNA interaction: evidence for nucleosome dissolution during replication and transcription and a low level of dissolution independent of both.Biochemistry. 1990 Jan 23;29(3):719-31. doi: 10.1021/bi00455a019. Biochemistry. 1990. PMID: 1692479
-
Mechanism of transcription through a nucleosome by RNA polymerase II.Biochim Biophys Acta. 2013 Jan;1829(1):76-83. doi: 10.1016/j.bbagrm.2012.08.015. Epub 2012 Sep 6. Biochim Biophys Acta. 2013. PMID: 22982194 Free PMC article. Review.
Cited by
-
Recombination-induced tag exchange (RITE) cassette series to monitor protein dynamics in Saccharomyces cerevisiae.G3 (Bethesda). 2013 Aug 7;3(8):1261-72. doi: 10.1534/g3.113.006213. G3 (Bethesda). 2013. PMID: 23708297 Free PMC article.
-
Spt5 histone binding activity preserves chromatin during transcription by RNA polymerase II.EMBO J. 2022 Mar 1;41(5):e109783. doi: 10.15252/embj.2021109783. Epub 2022 Feb 1. EMBO J. 2022. PMID: 35102600 Free PMC article.
-
Usage of the H3 variants during the S-phase of the cell cycle in Physarum polycephalum.Nucleic Acids Res. 2022 Mar 21;50(5):2536-2548. doi: 10.1093/nar/gkac060. Nucleic Acids Res. 2022. PMID: 35137186 Free PMC article.
-
Structure and function of the histone chaperone FACT - Resolving FACTual issues.Biochim Biophys Acta Gene Regul Mech. 2018 Jul 25:S1874-9399(18)30159-7. doi: 10.1016/j.bbagrm.2018.07.008. Online ahead of print. Biochim Biophys Acta Gene Regul Mech. 2018. PMID: 30055319 Free PMC article. Review.
-
Nucleosome retention by histone chaperones and remodelers occludes pervasive DNA-protein binding.Nucleic Acids Res. 2023 Sep 8;51(16):8496-8513. doi: 10.1093/nar/gkad615. Nucleic Acids Res. 2023. PMID: 37493599 Free PMC article.
References
-
- Ahmad K. and Henikoff, S. 2002. The histone variant H3.3 marks active chromatin by replication-independent nucleosome assembly. Mol. Cell 9: 1191-1200. - PubMed
-
- Baer B.W. and Rhodes, D. 1983. Eukaryotic RNA polymerase II binds to nucleosome cores from transcribed genes. Nature 301: 482-488. - PubMed
-
- Belotserkovskaya R., Oh, S., Bondarenko, V.A., Orphanides, G., Studitsky, V.M., and Reinberg, D. 2003. FACT facilitates transcription-dependent nucleosome alteration. Science 301: 1090-1093. - PubMed
-
- Boeger H., Griesenbeck, J., Strattan, J.S., and Kornberg, R.D. 2003. Nucleosomes unfold completely at a transcriptionally active promoter. Mol. Cell 11: 1587-1598. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
