Two distinct E3 ubiquitin ligases have complementary functions in the regulation of delta and serrate signaling in Drosophila
- PMID: 15760269
- PMCID: PMC1064853
- DOI: 10.1371/journal.pbio.0030096
Two distinct E3 ubiquitin ligases have complementary functions in the regulation of delta and serrate signaling in Drosophila
Abstract
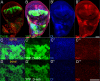
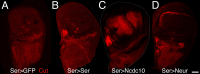
Signaling by the Notch ligands Delta (Dl) and Serrate (Ser) regulates a wide variety of essential cell-fate decisions during animal development. Two distinct E3 ubiquitin ligases, Neuralized (Neur) and Mind bomb (Mib), have been shown to regulate Dl signaling in Drosophila melanogaster and Danio rerio, respectively. While the neur and mib genes are evolutionarily conserved, their respective roles in the context of a single organism have not yet been examined. We show here that the Drosophila mind bomb (D-mib) gene regulates a subset of Notch signaling events, including wing margin specification, leg segmentation, and vein determination, that are distinct from those events requiring neur activity. D-mib also modulates lateral inhibition, a neur- and Dl-dependent signaling event, suggesting that D-mib regulates Dl signaling. During wing development, expression of D-mib in dorsal cells appears to be necessary and sufficient for wing margin specification, indicating that D-mib also regulates Ser signaling. Moreover, the activity of the D-mib gene is required for the endocytosis of Ser in wing imaginal disc cells. Finally, ectopic expression of neur in D-mib mutant larvae rescues the wing D-mib phenotype, indicating that Neur can compensate for the lack of D-mib activity. We conclude that D-mib and Neur are two structurally distinct proteins that have similar molecular activities but distinct developmental functions in Drosophila.
Figures


Similar articles
-
The ubiquitin ligase Drosophila Mind bomb promotes Notch signaling by regulating the localization and activity of Serrate and Delta.Development. 2005 May;132(10):2319-32. doi: 10.1242/dev.01825. Epub 2005 Apr 13. Development. 2005. PMID: 15829515
-
Distinct roles for Mind bomb, Neuralized and Epsin in mediating DSL endocytosis and signaling in Drosophila.Development. 2005 Jun;132(12):2883-94. doi: 10.1242/dev.01860. Development. 2005. PMID: 15930117
-
Functional analysis of the NHR2 domain indicates that oligomerization of Neuralized regulates ubiquitination and endocytosis of Delta during Notch signaling.Mol Cell Biol. 2012 Dec;32(24):4933-45. doi: 10.1128/MCB.00711-12. Epub 2012 Oct 8. Mol Cell Biol. 2012. PMID: 23045391 Free PMC article.
-
Delivering the lateral inhibition punchline: it's all about the timing.Sci Signal. 2010 Oct 26;3(145):pe38. doi: 10.1126/scisignal.3145pe38. Sci Signal. 2010. PMID: 20978236 Review.
-
Multiple levels of Notch signal regulation (review).Mol Membr Biol. 2002 Jan-Mar;19(1):27-38. doi: 10.1080/09687680110112929. Mol Membr Biol. 2002. PMID: 11989820 Review.
Cited by
-
Disruption of zebrafish cyclin G-associated kinase (GAK) function impairs the expression of Notch-dependent genes during neurogenesis and causes defects in neuronal development.BMC Dev Biol. 2010 Jan 18;10:7. doi: 10.1186/1471-213X-10-7. BMC Dev Biol. 2010. PMID: 20082716 Free PMC article.
-
Neuronal morphogenesis is regulated by the interplay between cyclin-dependent kinase 5 and the ubiquitin ligase mind bomb 1.J Neurosci. 2007 Aug 29;27(35):9503-12. doi: 10.1523/JNEUROSCI.1408-07.2007. J Neurosci. 2007. PMID: 17728463 Free PMC article.
-
The E3 ubiquitin ligase mindbomb1 controls planar cell polarity-dependent convergent extension movements during zebrafish gastrulation.Elife. 2022 Feb 10;11:e71928. doi: 10.7554/eLife.71928. Elife. 2022. PMID: 35142609 Free PMC article.
-
Regulation of EGFR and Notch signaling by distinct isoforms of D-cbl during Drosophila development.Dev Biol. 2010 Jun 1;342(1):1-10. doi: 10.1016/j.ydbio.2010.03.005. Epub 2010 Mar 17. Dev Biol. 2010. PMID: 20302857 Free PMC article.
-
Notch ligand ubiquitylation: what is it good for?Dev Cell. 2011 Jul 19;21(1):134-44. doi: 10.1016/j.devcel.2011.06.006. Dev Cell. 2011. PMID: 21763614 Free PMC article. Review.
References
-
- Lai EC. Notch signaling: Control of cell communication and cell fate. Development. 2004;131:965–973. - PubMed
-
- Schweisguth F. Notch signaling activity. Curr Biol. 2004;14:R129–R138. - PubMed
-
- Fleming RJ. Structural conservation of Notch receptors and ligands. Semin Cell Dev Biol. 1998;9:599–607. - PubMed
-
- Lehmann R, Jimenez F, Dietrich U, Campos-Ortega JA. On the phenotype and development of mutants of early neurogenesis in Drosophila melanogaster . Rouxs Arch Dev Biol. 1983;192:62–74. - PubMed
-
- Wiellette EL, McGinnis W. Hox genes differentially regulate Serrate to generate segment-specific structures. Development. 1999;126:1985–1995. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

