A configuration space of homologous proteins conserving mutual information and allowing a phylogeny inference based on pair-wise Z-score probabilities
- PMID: 15757521
- PMCID: PMC555736
- DOI: 10.1186/1471-2105-6-49
A configuration space of homologous proteins conserving mutual information and allowing a phylogeny inference based on pair-wise Z-score probabilities
Abstract
Background: Popular methods to reconstruct molecular phylogenies are based on multiple sequence alignments, in which addition or removal of data may change the resulting tree topology. We have sought a representation of homologous proteins that would conserve the information of pair-wise sequence alignments, respect probabilistic properties of Z-scores (Monte Carlo methods applied to pair-wise comparisons) and be the basis for a novel method of consistent and stable phylogenetic reconstruction.
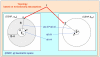
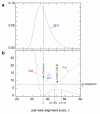
Results: We have built up a spatial representation of protein sequences using concepts from particle physics (configuration space) and respecting a frame of constraints deduced from pair-wise alignment score properties in information theory. The obtained configuration space of homologous proteins (CSHP) allows the representation of real and shuffled sequences, and thereupon an expression of the TULIP theorem for Z-score probabilities. Based on the CSHP, we propose a phylogeny reconstruction using Z-scores. Deduced trees, called TULIP trees, are consistent with multiple-alignment based trees. Furthermore, the TULIP tree reconstruction method provides a solution for some previously reported incongruent results, such as the apicomplexan enolase phylogeny.
Conclusion: The CSHP is a unified model that conserves mutual information between proteins in the way physical models conserve energy. Applications include the reconstruction of evolutionary consistent and robust trees, the topology of which is based on a spatial representation that is not reordered after addition or removal of sequences. The CSHP and its assigned phylogenetic topology, provide a powerful and easily updated representation for massive pair-wise genome comparisons based on Z-score computations.
Figures



Similar articles
-
Bayesian coestimation of phylogeny and sequence alignment.BMC Bioinformatics. 2005 Apr 1;6:83. doi: 10.1186/1471-2105-6-83. BMC Bioinformatics. 2005. PMID: 15804354 Free PMC article.
-
Scoredist: a simple and robust protein sequence distance estimator.BMC Bioinformatics. 2005 Apr 27;6:108. doi: 10.1186/1471-2105-6-108. BMC Bioinformatics. 2005. PMID: 15857510 Free PMC article.
-
Vestige: maximum likelihood phylogenetic footprinting.BMC Bioinformatics. 2005 May 29;6:130. doi: 10.1186/1471-2105-6-130. BMC Bioinformatics. 2005. PMID: 15921531 Free PMC article.
-
Differences between pair-wise and multi-sequence alignment methods affect vertebrate genome comparisons.Trends Genet. 2006 Apr;22(4):187-93. doi: 10.1016/j.tig.2006.02.005. Epub 2006 Feb 24. Trends Genet. 2006. PMID: 16499991 Review.
-
Multiple sequence alignment: in pursuit of homologous DNA positions.Genome Res. 2007 Feb;17(2):127-35. doi: 10.1101/gr.5232407. Genome Res. 2007. PMID: 17272647 Review.
Cited by
-
Evolutionary history expands the range of signaling interactions in hybrid multikinase networks.Sci Rep. 2021 Jun 3;11(1):11763. doi: 10.1038/s41598-021-91260-w. Sci Rep. 2021. PMID: 34083699 Free PMC article.
-
A simple derivation of the distribution of pairwise local protein sequence alignment scores.Evol Bioinform Online. 2008 Feb 14;4:41-5. Evol Bioinform Online. 2008. PMID: 19204806 Free PMC article.
-
Amino acid "little Big Bang": representing amino acid substitution matrices as dot products of Euclidian vectors.BMC Bioinformatics. 2010 Jan 4;11:4. doi: 10.1186/1471-2105-11-4. BMC Bioinformatics. 2010. PMID: 20047649 Free PMC article.
-
Evolution of biological sequences implies an extreme value distribution of type I for both global and local pairwise alignment scores.BMC Bioinformatics. 2008 Aug 7;9:332. doi: 10.1186/1471-2105-9-332. BMC Bioinformatics. 2008. PMID: 18687111 Free PMC article.
-
Evolution of folate biosynthesis and metabolism across algae and land plant lineages.Sci Rep. 2019 Apr 5;9(1):5731. doi: 10.1038/s41598-019-42146-5. Sci Rep. 2019. PMID: 30952916 Free PMC article.
References
-
- Zuckerkandl E, Pauling L. Molecules as documents of evolutionary history. J Theor Biol. 1965;8:357–366. - PubMed
-
- Zukerkandl E. The evolution of hemoglobin. Sci Am. 1965;212:110–118. - PubMed
-
- Fitch WM, Margoliash E. Construction of phylogenetic trees. Science. 1967;155:279–284. - PubMed
-
- Arnheim N, Taylor CE. Non-Darwinian evolution: consequences for neutral allelic variation. Nature. 1969;223:900–903. - PubMed
-
- Dayhoff MO. Computer analysis of protein evolution. Sci Am. 1969;221:86–95. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

