Transcriptome analysis of the Rhodobacter sphaeroides PpsR regulon: PpsR as a master regulator of photosystem development
- PMID: 15743963
- PMCID: PMC1064034
- DOI: 10.1128/JB.187.6.2148-2156.2005
Transcriptome analysis of the Rhodobacter sphaeroides PpsR regulon: PpsR as a master regulator of photosystem development
Abstract
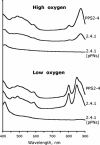
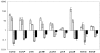
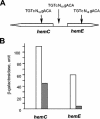
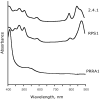
PpsR from the anoxygenic phototrophic bacterium Rhodobacter sphaeroides has been known as an oxygen- and light-dependent repressor of bacteriochlorophyll and carotenoid biosynthesis genes and puc operons involved in photosystem development. However, the putative PpsR-binding sites, TGTN12ACA, are also located upstream of numerous nonphotosystem genes, thus raising the possibility that the role of PpsR is broader. To characterize the PpsR regulon, transcriptome profiling was performed on the wild-type strain grown at high and low oxygen tensions, on the strain overproducing PpsR, and on the ppsR mutant. Transcriptome analysis showed that PpsR primarily regulates photosystem genes; the consensus PpsR binding sequence is TGTcN10gACA (lowercase letters indicate lesser conservation); the presence of two binding sites is required for repression in vivo. These findings explain why numerous single TGTN12ACA sequences are nonfunctional. In addition to photosystem genes, the hemC and hemE genes involved in the early steps of tetrapyrrole biosynthesis were identified as new direct targets of PpsR repression. Unexpectedly, PpsR was found to indirectly repress the puf and puhA operons encoding photosystem core proteins. The upstream regions of these operons contain no PpsR binding sites. Involvement in regulation of these operons suggests that PpsR functions as a master regulator of photosystem development. Upregulation of the puf and puhA operons that resulted from ppsR inactivation was sufficient to restore the ability to grow phototrophically to the prrA mutant. PrrA, the global redox-dependent activator, was previously considered indispensable for phototrophic growth. It is revealed that the PrrBA and AppA-PpsR systems, believed to work independently, in fact interact and coordinately regulate photosystem development.
Figures



Similar articles
-
The AppA and PpsR proteins from Rhodobacter sphaeroides can establish a redox-dependent signal chain but fail to transmit blue-light signals in other bacteria.J Bacteriol. 2007 Mar;189(6):2274-82. doi: 10.1128/JB.01699-06. Epub 2007 Jan 5. J Bacteriol. 2007. PMID: 17209035 Free PMC article.
-
Hierarchical regulation of photosynthesis gene expression by the oxygen-responsive PrrBA and AppA-PpsR systems of Rhodobacter sphaeroides.J Bacteriol. 2008 Dec;190(24):8106-14. doi: 10.1128/JB.01094-08. Epub 2008 Oct 17. J Bacteriol. 2008. PMID: 18931128 Free PMC article.
-
Molecular genetic analysis suggesting interactions between AppA and PpsR in regulation of photosynthesis gene expression in Rhodobacter sphaeroides 2.4.1.J Bacteriol. 1997 Jan;179(1):128-34. doi: 10.1128/jb.179.1.128-134.1997. J Bacteriol. 1997. PMID: 8981989 Free PMC article.
-
PpsR: a multifaceted regulator of photosynthesis gene expression in purple bacteria.Mol Microbiol. 2005 Jul;57(1):17-26. doi: 10.1111/j.1365-2958.2005.04655.x. Mol Microbiol. 2005. PMID: 15948946 Review.
-
The PpsR regulator family.Res Microbiol. 2005 Jun-Jul;156(5-6):619-25. doi: 10.1016/j.resmic.2005.02.001. Epub 2005 Feb 25. Res Microbiol. 2005. PMID: 15950121 Review.
Cited by
-
Reconstruction of the core and extended regulons of global transcription factors.PLoS Genet. 2010 Jul 22;6(7):e1001027. doi: 10.1371/journal.pgen.1001027. PLoS Genet. 2010. PMID: 20661434 Free PMC article.
-
The complete genome sequence of Roseobacter denitrificans reveals a mixotrophic rather than photosynthetic metabolism.J Bacteriol. 2007 Feb;189(3):683-90. doi: 10.1128/JB.01390-06. Epub 2006 Nov 10. J Bacteriol. 2007. PMID: 17098896 Free PMC article.
-
Multi-PAS domain-mediated protein oligomerization of PpsR from Rhodobacter sphaeroides.Acta Crystallogr D Biol Crystallogr. 2014 Mar;70(Pt 3):863-76. doi: 10.1107/S1399004713033634. Epub 2014 Feb 27. Acta Crystallogr D Biol Crystallogr. 2014. PMID: 24598755 Free PMC article.
-
The AppA and PpsR proteins from Rhodobacter sphaeroides can establish a redox-dependent signal chain but fail to transmit blue-light signals in other bacteria.J Bacteriol. 2007 Mar;189(6):2274-82. doi: 10.1128/JB.01699-06. Epub 2007 Jan 5. J Bacteriol. 2007. PMID: 17209035 Free PMC article.
-
Bacteriophytochromes in anoxygenic photosynthetic bacteria.Photosynth Res. 2008 Aug;97(2):141-53. doi: 10.1007/s11120-008-9323-0. Epub 2008 Jul 9. Photosynth Res. 2008. PMID: 18612842 Review.
References
-
- Bauer, C. E., J. J. Buggy, Z. M. Yang, and B. L. Marrs. 1991. The superoperonal organization of genes for pigment biosynthesis and reaction center proteins is a conserved feature in Rhodobacter capsulatus: analysis of overlapping bchB and puhA transcripts. Mol. Gen. Genet. 228:443-454. - PubMed
-
- Braatsch, S., M. Gomelsky, S. Kuphal, and G. Klug. 2002. The single flavoprotein, AppA, from Rhodobacter sphaeroides integrates both redox and light signals. Mol. Microbiol. 45:827-836. - PubMed
-
- Cho, S. H., S. H. Youn, S. R. Lee, H. S. Yim, and S. O. Kang. 2004. Redox property and regulation of PpsR, a transcriptional repressor of photosystem gene expression in Rhodobacter sphaeroides. Microbiology 150:697-706. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

