Identification and dynamics of two classes of aurora-like kinases in Arabidopsis and other plants
- PMID: 15722465
- PMCID: PMC1069702
- DOI: 10.1105/tpc.104.029710
Identification and dynamics of two classes of aurora-like kinases in Arabidopsis and other plants
Abstract
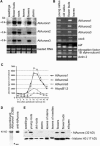
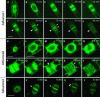
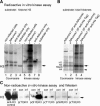
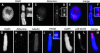
Aurora-like kinases play key roles in chromosome segregation and cytokinesis in yeast, plant, and animal systems. Here, we characterize three Arabidopsis thaliana protein kinases, designated AtAurora1, AtAurora2, and AtAurora3, which share high amino acid identities with the Ser/Thr kinase domain of yeast Ipl1 and animal Auroras. Structure and expression of AtAurora1 and AtAurora2 suggest that these genes arose by a recent gene duplication, whereas the diversification of plant alpha and beta Aurora kinases predates the origin of land plants. The transcripts and proteins of all three kinases are most abundant in tissues containing dividing cells. Intracellular localization of green fluorescent protein-tagged AtAuroras revealed an AtAurora-type specific association mainly with dynamic mitotic structures, such as microtubule spindles and centromeres, and with the emerging cell plate of dividing tobacco (Nicotiana tabacum) BY-2 cells. Immunolabeling using AtAurora antibodies yielded specific signals at the centromeres that are coincident with histone H3 that is phosphorylated at Ser position10 during mitosis. An in vitro kinase assay demonstrated that AtAurora1 preferentially phosphorylates histone H3 at Ser 10 but not at Ser 28 or Thr 3, 11, and 32. The phylogenetic analysis of available Aurora sequences from different eukaryotic origins suggests that, although a plant Aurora gene has been duplicated early in the evolution of plants, the paralogs nevertheless maintained a role in cell cycle-related signal transduction pathways.
Figures


Similar articles
-
Characterization of plant Aurora kinases during mitosis.Plant Mol Biol. 2005 May;58(1):1-13. doi: 10.1007/s11103-005-3454-x. Plant Mol Biol. 2005. PMID: 16028112
-
Identification and characterization of plant Haspin kinase as a histone H3 threonine kinase.BMC Plant Biol. 2011 Apr 28;11:73. doi: 10.1186/1471-2229-11-73. BMC Plant Biol. 2011. PMID: 21527018 Free PMC article.
-
Altered expression of Aurora kinases in Arabidopsis results in aneu- and polyploidization.Plant J. 2014 Nov;80(3):449-61. doi: 10.1111/tpj.12647. Epub 2014 Sep 19. Plant J. 2014. PMID: 25146886
-
Phosphorylation of histone H3 in plants--a dynamic affair.Biochim Biophys Acta. 2007 May-Jun;1769(5-6):308-15. doi: 10.1016/j.bbaexp.2007.01.002. Epub 2007 Jan 19. Biochim Biophys Acta. 2007. PMID: 17320987 Review.
-
The phosphoinositide-3-OH-kinase-related kinases of Arabidopsis thaliana.EMBO Rep. 2005 Aug;6(8):723-8. doi: 10.1038/sj.embor.7400479. EMBO Rep. 2005. PMID: 16065066 Free PMC article. Review.
Cited by
-
Cyanidioschyzon merolae aurora kinase phosphorylates evolutionarily conserved sites on its target to regulate mitochondrial division.Commun Biol. 2019 Dec 20;2:477. doi: 10.1038/s42003-019-0714-x. eCollection 2019. Commun Biol. 2019. PMID: 31886415 Free PMC article.
-
Phosphorylation of Plant Microtubule-Associated Proteins During Cell Division.Front Plant Sci. 2019 Mar 11;10:238. doi: 10.3389/fpls.2019.00238. eCollection 2019. Front Plant Sci. 2019. PMID: 30915087 Free PMC article. Review.
-
The plant TPX2 protein regulates prospindle assembly before nuclear envelope breakdown.Plant Cell. 2008 Oct;20(10):2783-97. doi: 10.1105/tpc.107.056796. Epub 2008 Oct 21. Plant Cell. 2008. PMID: 18941054 Free PMC article.
-
Random chromosome distribution in the first meiosis of F1 disomic substitution line 2R(2D) x rye hybrids (ABDR, 4× = 28) occurs without bipolar spindle assembly.Comp Cytogenet. 2020 Oct 9;14(4):453-482. doi: 10.3897/compcytogen.v14.i4.55827. eCollection 2020. Comp Cytogenet. 2020. PMID: 33117496 Free PMC article.
-
Functional Divergence of Microtubule-Associated TPX2 Family Members in Arabidopsis thaliana.Int J Mol Sci. 2020 Mar 22;21(6):2183. doi: 10.3390/ijms21062183. Int J Mol Sci. 2020. PMID: 32235723 Free PMC article.
References
-
- Adams, R.R., Carmena, M., and Earnshaw, W.C. (2001. a). Chromosomal passengers and the (aurora) ABCs of mitosis. Trends Cell Biol. 11, 49–54. - PubMed
-
- Adams, R.R., Carmena, M., and Earnshaw, W.C. (2001. b). Chromosomal passengers and the (aurora) ABCs of mitosis. Trends Cell Biol. 11, 49–54. - PubMed
-
- Adams, R.R., Maiato, H., Earnshaw, W.C., and Carmena, M. (2001. c). Essential roles of Drosophila inner centromere protein (INCENP) and aurora B in histone H3 phosphorylation, metaphase chromosome alignment, kinetochore disjunction, and chromosome segregation. J. Cell Biol. 153, 865–880. - PMC - PubMed
-
- Andrews, P.D., Knatko, E., Moore, W.J., and Swedlow, J.R. (2003). Mitotic mechanics: The auroras come into view. Curr. Opin. Cell Biol. 15, 672–683. - PubMed
-
- Arlot-Bonnemains, Y., Klotzbucher, A., Giet, R., Uzbekov, R., Bihan, R., and Prigent, C. (2001). Identification of a functional destruction box in the Xenopus laevis aurora-A kinase pEg2. FEBS Lett. 508, 149–152. - PubMed
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

