Methylation as a crucial step in plant microRNA biogenesis
- PMID: 15705854
- PMCID: PMC5137370
- DOI: 10.1126/science.1107130
Methylation as a crucial step in plant microRNA biogenesis
Abstract
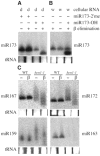
Methylation on the base or the ribose is prevalent in eukaryotic ribosomal RNAs (rRNAs) and is thought to be crucial for ribosome biogenesis and function. Artificially introduced 2'-O-methyl groups in small interfering RNAs (siRNAs) can stabilize siRNAs in serum without affecting their activities in RNA interference in mammalian cells. Here, we show that plant microRNAs (miRNAs) have a naturally occurring methyl group on the ribose of the last nucleotide. Whereas methylation of rRNAs depends on guide RNAs, the methyltransferase protein HEN1 is sufficient to methylate miRNA/miRNA* duplexes. Our studies uncover a new and crucial step in plant miRNA biogenesis and have profound implications in the function of miRNAs.
Figures


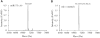
Similar articles
-
Kinetic and functional analysis of the small RNA methyltransferase HEN1: the catalytic domain is essential for preferential modification of duplex RNA.RNA. 2010 Oct;16(10):1935-42. doi: 10.1261/rna.2281410. Epub 2010 Aug 12. RNA. 2010. PMID: 20705645 Free PMC article.
-
MicroRNA biogenesis and function in plants.FEBS Lett. 2005 Oct 31;579(26):5923-31. doi: 10.1016/j.febslet.2005.07.071. Epub 2005 Aug 9. FEBS Lett. 2005. PMID: 16144699 Free PMC article. Review.
-
Degradation of microRNAs by a family of exoribonucleases in Arabidopsis.Science. 2008 Sep 12;321(5895):1490-2. doi: 10.1126/science.1163728. Science. 2008. PMID: 18787168 Free PMC article.
-
siRNAs compete with miRNAs for methylation by HEN1 in Arabidopsis.Nucleic Acids Res. 2010 Sep;38(17):5844-50. doi: 10.1093/nar/gkq348. Epub 2010 May 6. Nucleic Acids Res. 2010. PMID: 20448024 Free PMC article.
-
The expanding world of small RNAs in plants.Nat Rev Mol Cell Biol. 2015 Dec;16(12):727-41. doi: 10.1038/nrm4085. Epub 2015 Nov 4. Nat Rev Mol Cell Biol. 2015. PMID: 26530390 Free PMC article. Review.
Cited by
-
Meta-2OM: A multi-classifier meta-model for the accurate prediction of RNA 2'-O-methylation sites in human RNA.PLoS One. 2024 Jun 26;19(6):e0305406. doi: 10.1371/journal.pone.0305406. eCollection 2024. PLoS One. 2024. PMID: 38924058 Free PMC article.
-
Uptake of MicroRNAs from Exosome-Like Nanovesicles of Edible Plant Juice by Rat Enterocytes.Int J Mol Sci. 2021 Apr 3;22(7):3749. doi: 10.3390/ijms22073749. Int J Mol Sci. 2021. PMID: 33916868 Free PMC article.
-
Characterization of microRNAs expression during maize seed development.BMC Genomics. 2012 Aug 1;13:360. doi: 10.1186/1471-2164-13-360. BMC Genomics. 2012. PMID: 22853295 Free PMC article.
-
Recent advances in the regulation of plant miRNA biogenesis.RNA Biol. 2021 Dec;18(12):2087-2096. doi: 10.1080/15476286.2021.1899491. Epub 2021 Mar 17. RNA Biol. 2021. PMID: 33666136 Free PMC article. Review.
-
MicroRNAs and drought responses in sugarcane.Front Plant Sci. 2015 Feb 23;6:58. doi: 10.3389/fpls.2015.00058. eCollection 2015. Front Plant Sci. 2015. PMID: 25755657 Free PMC article. Review.
References
-
- Bartel DP. Cell. 2004;116:281. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

