Solution structure of the PWWP domain of the hepatoma-derived growth factor family
- PMID: 15689505
- PMCID: PMC2279273
- DOI: 10.1110/ps.04975305
Solution structure of the PWWP domain of the hepatoma-derived growth factor family
Abstract
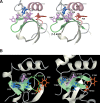
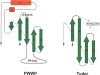
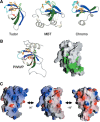
Among the many PWWP-containing proteins, the largest group of homologous proteins is related to hepatoma-derived growth factor (HDGF). Within a well-conserved region at the extreme N-terminus, HDGF and five HDGF-related proteins (HRPs) always have a PWWP domain, which is a module found in many chromatin-associated proteins. In this study, we determined the solution structure of the PWWP domain of HDGF-related protein-3 (HRP-3) by NMR spectroscopy. The structure consists of a five-stranded beta-barrel with a PWWP-specific long loop connecting beta2 and beta3 (PR-loop), followed by a helical region including two alpha-helices. Its structure was found to have a characteristic solvent-exposed hydrophobic cavity, which is composed of an abundance of aromatic residues in the beta1/beta2 loop (beta-beta arch) and the beta3/beta4 loop. A similar ligand binding cavity occurs at the corresponding position in the Tudor, chromo, and MBT domains, which have structural and probable evolutionary relationships with PWWP domains. These findings suggest that the PWWP domains of the HDGF family bind to some component of chromatin via the cavity.
Figures

Similar articles
-
High resolution structure of the HDGF PWWP domain: a potential DNA binding domain.Protein Sci. 2006 Feb;15(2):314-23. doi: 10.1110/ps.051751706. Epub 2005 Dec 29. Protein Sci. 2006. PMID: 16384999 Free PMC article.
-
Domain swapping and SMYD1 interactions with the PWWP domain of human hepatoma-derived growth factor.Sci Rep. 2018 Jan 10;8(1):287. doi: 10.1038/s41598-017-18510-8. Sci Rep. 2018. PMID: 29321480 Free PMC article.
-
The First Residue of the PWWP Motif Modulates HATH Domain Binding, Stability, and Protein-Protein Interaction.Biochemistry. 2015 Jul 7;54(26):4063-74. doi: 10.1021/acs.biochem.5b00454. Epub 2015 Jun 24. Biochemistry. 2015. PMID: 26067205
-
The PWWP domain: a potential protein-protein interaction domain in nuclear proteins influencing differentiation?FEBS Lett. 2000 May 4;473(1):1-5. doi: 10.1016/s0014-5793(00)01449-6. FEBS Lett. 2000. PMID: 10802047 Review.
-
Structure and function of the nucleosome-binding PWWP domain.Trends Biochem Sci. 2014 Nov;39(11):536-47. doi: 10.1016/j.tibs.2014.09.001. Epub 2014 Sep 29. Trends Biochem Sci. 2014. PMID: 25277115 Review.
Cited by
-
Keeping it in the family: diverse histone recognition by conserved structural folds.Crit Rev Biochem Mol Biol. 2010 Dec;45(6):488-505. doi: 10.3109/10409238.2010.512001. Epub 2010 Oct 6. Crit Rev Biochem Mol Biol. 2010. PMID: 20923397 Free PMC article. Review.
-
Role of the PWWP domain of lens epithelium-derived growth factor (LEDGF)/p75 cofactor in lentiviral integration targeting.J Biol Chem. 2011 Dec 2;286(48):41812-41826. doi: 10.1074/jbc.M111.255711. Epub 2011 Oct 10. J Biol Chem. 2011. Retraction in: J Biol Chem. 2018 Jan 5;293(1):114. doi: 10.1074/jbc.W117.001458. PMID: 21987578 Free PMC article. Retracted.
-
CCNI2 promotes the progression of human gastric cancer through HDGF.Cancer Cell Int. 2021 Dec 11;21(1):661. doi: 10.1186/s12935-021-02352-6. Cancer Cell Int. 2021. PMID: 34895232 Free PMC article.
-
[Aberrant expression of HDGF and its prognostic values in surgically resected non-small cell lung cancer].Zhongguo Fei Ai Za Zhi. 2011 Mar;14(3):211-8. doi: 10.3779/j.issn.1009-3419.2011.03.06. Zhongguo Fei Ai Za Zhi. 2011. PMID: 21426662 Free PMC article. Chinese.
-
Structural basis for high-affinity binding of LEDGF PWWP to mononucleosomes.Nucleic Acids Res. 2013 Apr 1;41(6):3924-36. doi: 10.1093/nar/gkt074. Epub 2013 Feb 8. Nucleic Acids Res. 2013. PMID: 23396443 Free PMC article.
References
-
- Abouzied, M.M., Baader, S.L., Dietz, F., Kappler, J., Gieselmann, V., and Franken, S. 2004. Expression patterns and different subcellular localization of the growth factors HDGF (hepatoma-derived growth factor) and HRP-3 (HDGF-related protein-3) suggest functions in addition to their mitogenic activity. Biochem. J. 378 169–176. - PMC - PubMed
-
- Bax, A. 1994. Multidimensional nuclear magnetic resonance methods for protein studies. Curr. Opin. Struct. Biol. 4 738–744.
-
- Cavanagh, J., Fairbrother, W.J., Palmer III, A.G., and Skelton, N.J. 1996. Protein NMR spectroscopy, principles and practice. Academic Press, San Diego, CA.
-
- Cornell, W.D., Cieplak, P., Bayly, C.I., Gould, I.R., Merz, K.M., Ferguson, D.M., Spellmeyer, D.C., Fox, T., Caldwell, J.W., and Kollman, P.A. 1995. A second generation force field for the simulation of proteins, nucleic acids, and organic molecules. J. Am. Chem. Soc. 117 5179–5197.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

