Role for SUMO modification in facilitating transcriptional repression by BKLF
- PMID: 15684403
- PMCID: PMC548027
- DOI: 10.1128/MCB.25.4.1549-1559.2005
Role for SUMO modification in facilitating transcriptional repression by BKLF
Abstract
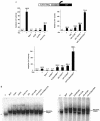
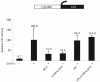
Small ubiquitin-like modifier (SUMO) is a protein moiety that is ligated to lysine residues on a variety of target proteins. Many known SUMO substrates are transcription factors or coregulators of transcription, and in most cases, modification with SUMO leads to the attenuation of transcriptional activation. We have examined basic Kruppel-like factor/Kruppel-like factor 3 (BKLF), a zinc finger transcription factor that is known to function as a potent transcriptional repressor. We show that BKLF recruits the E2 SUMO-conjugating enzyme Ubc9 and can be modified by the addition of SUMO-1 in vitro and in vivo. The SUMO E3 ligases PIAS1, PIASgamma, PIASxalpha, and PIASxbeta but not Pc2 enhance the sumoylation of BKLF. Site-directed mutagenesis identified two lysines (K10 and K197) of BKLF as the sumoylation sites. Sumoylation does not detectably affect DNA binding by BKLF, but mutation of the sumoylation sites reduces transcriptional repression activity. Most interestingly, when mutations preventing sumoylation are combined with an additional mutation that eliminates contact with the C-terminal binding protein (CtBP) corepressor, BKLF becomes an activator of transcription. These results link SUMO modification to transcriptional repression and demonstrate that both recruitment of CtBP and sumoylation are required for full repression by BKLF.
Figures

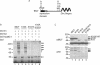


Similar articles
-
The transcriptional repression activity of KyoT2 on the Notch/RBP-J pathway is regulated by PIAS1-catalyzed SUMOylation.J Mol Biol. 2007 Jun 29;370(1):27-38. doi: 10.1016/j.jmb.2007.04.010. Epub 2007 Apr 12. J Mol Biol. 2007. PMID: 17509614
-
SUMO modification modulates the transrepression activity of PLZF.Biochem Biophys Res Commun. 2007 Jun 29;358(2):475-82. doi: 10.1016/j.bbrc.2007.04.157. Epub 2007 May 4. Biochem Biophys Res Commun. 2007. PMID: 17498654
-
Sumoylation of CoREST modulates its function as a transcriptional repressor.Biochem Biophys Res Commun. 2008 Dec 26;377(4):1031-5. doi: 10.1016/j.bbrc.2008.09.149. Epub 2008 Oct 12. Biochem Biophys Res Commun. 2008. PMID: 18854179
-
Transcriptional regulation by C-terminal binding proteins.Int J Biochem Cell Biol. 2007;39(9):1593-607. doi: 10.1016/j.biocel.2007.01.025. Epub 2007 Feb 4. Int J Biochem Cell Biol. 2007. PMID: 17336131 Review.
-
Basic Krüppel-like factor functions within a network of interacting haematopoietic transcription factors.Int J Biochem Cell Biol. 1999 Oct;31(10):1169-74. doi: 10.1016/s1357-2725(99)00067-9. Int J Biochem Cell Biol. 1999. PMID: 10582345 Review.
Cited by
-
Arabidopsis HFR1 is a potential nuclear substrate regulated by the Xanthomonas type III effector XopD(Xcc8004).PLoS One. 2015 Feb 3;10(2):e0117067. doi: 10.1371/journal.pone.0117067. eCollection 2015. PLoS One. 2015. PMID: 25647296 Free PMC article.
-
Epithelial sodium channel (ENaC) is multi-ubiquitinated at the cell surface.Biochem J. 2007 Jul 1;405(1):147-55. doi: 10.1042/BJ20060747. Biochem J. 2007. PMID: 17381423 Free PMC article.
-
A small ubiquitin-related modifier-interacting motif functions as the transcriptional activation domain of Krüppel-like factor 4.J Biol Chem. 2010 Sep 3;285(36):28298-308. doi: 10.1074/jbc.M110.101717. Epub 2010 Jun 28. J Biol Chem. 2010. PMID: 20584900 Free PMC article.
-
MiR-326/Sp1/KLF3: A novel regulatory axis in lung cancer progression.Cell Prolif. 2019 Mar;52(2):e12551. doi: 10.1111/cpr.12551. Epub 2018 Nov 28. Cell Prolif. 2019. PMID: 30485570 Free PMC article.
-
CRMP2 protein SUMOylation modulates NaV1.7 channel trafficking.J Biol Chem. 2013 Aug 23;288(34):24316-31. doi: 10.1074/jbc.M113.474924. Epub 2013 Jul 8. J Biol Chem. 2013. PMID: 23836888 Free PMC article.
References
-
- Abdel-Hafiz, H. A., G. S. Takimoto, L. Tung, and K. B. Horwitz. 2002. The inhibitory function (IF) in human progesterone receptor N-termini binds small ubiquitin-like modifier (SUMO-1) protein to regulate autoinhibition and transrepression. J. Biol. Chem. 277:33950-33956. - PubMed
-
- Bies, J., J. Markus, and L. Wolff. 2002. Covalent attachment of the SUMO-1 protein to the negative regulatory domain of the c-Myb transcription factor modifies its stability and transactivation capacity. J. Biol. Chem. 277:8999-9009. - PubMed
-
- Courey, A. J., and R. Tjian. 1988. Analysis of Sp1 in vivo reveals multiple transcriptional domains, including a novel glutamine-rich activation motif. Cell 55:887-898. - PubMed
-
- David, G., M. A. Neptune, and R. A. DePinho. 2002. SUMO-1 modification of histone deacetylase 1 (HDAC1) modulates its biological activities. J. Biol. Chem. 277:23658-23663. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous
